Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
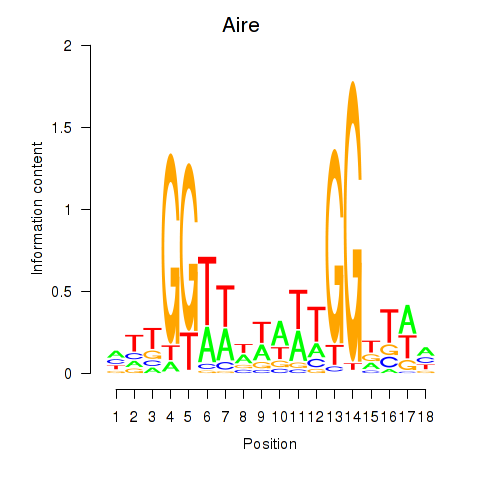
Results for Aire
Z-value: 0.96
Transcription factors associated with Aire
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Aire
|
ENSRNOG00000001213 | autoimmune regulator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Aire | rn6_v1_chr20_+_11365697_11365697 | -0.26 | 2.1e-06 | Click! |
Activity profile of Aire motif
Sorted Z-values of Aire motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_200762206 | 35.99 |
ENSRNOT00000068511
ENSRNOT00000086835 |
Hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chrM_-_14061 | 31.46 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chrM_+_14136 | 27.24 |
ENSRNOT00000042098
|
Mt-cyb
|
mitochondrially encoded cytochrome b |
| chr2_-_200762492 | 26.29 |
ENSRNOT00000056172
|
Hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr3_+_128155069 | 24.88 |
ENSRNOT00000051184
ENSRNOT00000006389 |
Plcb1
|
phospholipase C beta 1 |
| chr16_-_79671719 | 24.74 |
ENSRNOT00000015908
|
Myom2
|
myomesin 2 |
| chrM_+_7006 | 24.55 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr7_+_48867664 | 21.85 |
ENSRNOT00000005862
|
Ppfia2
|
PTPRF interacting protein alpha 2 |
| chr1_+_101884019 | 18.41 |
ENSRNOT00000028650
|
Tmem143
|
transmembrane protein 143 |
| chr11_-_31772984 | 17.51 |
ENSRNOT00000002771
|
Dnajc28
|
DnaJ heat shock protein family (Hsp40) member C28 |
| chr9_+_12475006 | 17.11 |
ENSRNOT00000079703
|
LOC100912293
|
uncharacterized LOC100912293 |
| chrM_+_9451 | 16.96 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr4_+_88695590 | 16.79 |
ENSRNOT00000087835
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr5_-_17061837 | 16.39 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chrM_+_7758 | 15.21 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr20_-_4823475 | 14.89 |
ENSRNOT00000082536
ENSRNOT00000001114 |
Atp6v1g2
|
ATPase H+ transporting V1 subunit G2 |
| chr2_-_179704629 | 14.79 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr2_+_83393282 | 14.73 |
ENSRNOT00000044871
|
Ctnnd2
|
catenin delta 2 |
| chr8_+_49418965 | 14.72 |
ENSRNOT00000021819
|
Scn2b
|
sodium voltage-gated channel beta subunit 2 |
| chr3_-_51612397 | 13.63 |
ENSRNOT00000081401
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chrM_+_8599 | 13.22 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chrM_+_11736 | 13.01 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr4_-_120559078 | 12.95 |
ENSRNOT00000085730
ENSRNOT00000079575 |
Kbtbd12
|
kelch repeat and BTB domain containing 12 |
| chr3_+_151335292 | 12.68 |
ENSRNOT00000073642
|
Mmp24
|
matrix metallopeptidase 24 |
| chr20_+_34633157 | 12.59 |
ENSRNOT00000000469
|
Pln
|
phospholamban |
| chr1_+_228142778 | 12.01 |
ENSRNOT00000028517
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr7_-_138483612 | 11.76 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr11_-_4332255 | 11.69 |
ENSRNOT00000087133
|
Cadm2
|
cell adhesion molecule 2 |
| chr1_+_101884276 | 11.56 |
ENSRNOT00000082917
|
Tmem143
|
transmembrane protein 143 |
| chr12_+_40417944 | 11.19 |
ENSRNOT00000073125
|
Acad10
|
acyl-CoA dehydrogenase family, member 10 |
| chr10_-_27862868 | 11.07 |
ENSRNOT00000004877
|
Gabra6
|
gamma-aminobutyric acid type A receptor alpha 6 subunit |
| chr1_+_59156251 | 10.97 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
| chr2_+_182006242 | 10.54 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr3_-_48831417 | 10.34 |
ENSRNOT00000009920
ENSRNOT00000085246 |
Kcnh7
|
potassium voltage-gated channel subfamily H member 7 |
| chr17_+_68477446 | 10.19 |
ENSRNOT00000086236
ENSRNOT00000068174 |
Pitrm1
|
pitrilysin metallopeptidase 1 |
| chr7_+_123482255 | 9.91 |
ENSRNOT00000064487
|
LOC688613
|
hypothetical protein LOC688613 |
| chr1_-_154165524 | 9.22 |
ENSRNOT00000023468
|
Hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chr4_-_23135354 | 9.18 |
ENSRNOT00000011432
|
Steap4
|
STEAP4 metalloreductase |
| chr9_-_105693357 | 9.14 |
ENSRNOT00000066968
|
Nudt12
|
nudix hydrolase 12 |
| chr1_+_266482858 | 9.10 |
ENSRNOT00000027223
|
As3mt
|
arsenite methyltransferase |
| chr5_+_60528997 | 9.09 |
ENSRNOT00000051445
|
Grhpr
|
glyoxylate and hydroxypyruvate reductase |
| chr2_-_210282352 | 8.76 |
ENSRNOT00000075653
|
Slc6a17
|
solute carrier family 6 member 17 |
| chr7_-_123655896 | 8.58 |
ENSRNOT00000012413
|
Cyp2d2
|
cytochrome P450, family 2, subfamily d, polypeptide 2 |
| chr4_-_179339795 | 8.32 |
ENSRNOT00000080410
|
Bcat1
|
branched chain amino acid transaminase 1 |
| chr17_-_84614228 | 8.30 |
ENSRNOT00000043042
|
AABR07028748.1
|
|
| chrX_+_26294066 | 8.07 |
ENSRNOT00000037862
|
Hccs
|
holocytochrome c synthase |
| chr4_+_155709613 | 7.97 |
ENSRNOT00000013272
|
Necap1
|
NECAP endocytosis associated 1 |
| chr1_-_146556171 | 7.80 |
ENSRNOT00000017636
|
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr2_-_41785792 | 7.65 |
ENSRNOT00000015871
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr16_-_64716224 | 7.52 |
ENSRNOT00000071166
|
Fut10
|
fucosyltransferase 10 |
| chr4_-_7582207 | 7.46 |
ENSRNOT00000013932
|
Klhl7
|
kelch-like family member 7 |
| chr3_+_77152425 | 7.24 |
ENSRNOT00000092045
ENSRNOT00000082255 |
Olr653
|
olfactory receptor 653 |
| chr1_-_72509431 | 6.94 |
ENSRNOT00000071971
|
LOC684270
|
similar to isochorismatase domain containing 2 |
| chr5_-_124403195 | 6.94 |
ENSRNOT00000067850
|
C8a
|
complement C8 alpha chain |
| chr10_+_37594578 | 6.92 |
ENSRNOT00000007676
|
Skp1
|
S-phase kinase-associated protein 1 |
| chr6_-_142385773 | 6.82 |
ENSRNOT00000071555
|
AABR07065814.7
|
|
| chr1_+_94793195 | 6.40 |
ENSRNOT00000079174
|
AC120712.2
|
|
| chr7_-_9711928 | 6.31 |
ENSRNOT00000011167
|
Neurod4
|
neuronal differentiation 4 |
| chr1_+_71844368 | 6.12 |
ENSRNOT00000044754
|
Vom1r32
|
vomeronasal 1 receptor 32 |
| chr11_+_64790801 | 5.99 |
ENSRNOT00000004023
|
Timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chrX_-_10218583 | 5.72 |
ENSRNOT00000013382
|
Nyx
|
nyctalopin |
| chr4_+_88423412 | 5.70 |
ENSRNOT00000040163
|
Vom1r88
|
vomeronasal 1 receptor 88 |
| chr3_-_113312127 | 5.65 |
ENSRNOT00000065285
|
Mfap1a
|
microfibrillar-associated protein 1A |
| chr3_+_95233874 | 5.63 |
ENSRNOT00000079743
|
LOC691083
|
hypothetical protein LOC691083 |
| chr10_-_52290657 | 5.54 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr3_-_153042395 | 5.50 |
ENSRNOT00000055232
|
Ndrg3
|
NDRG family member 3 |
| chr2_-_21931720 | 5.15 |
ENSRNOT00000018449
|
Msh3
|
mutS homolog 3 |
| chr7_+_44009069 | 5.04 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr20_+_22728208 | 4.98 |
ENSRNOT00000000794
|
LOC100363472
|
nuclear receptor-binding factor 2-like |
| chr4_+_88184956 | 4.96 |
ENSRNOT00000077129
|
Vom1r83
|
vomeronasal 1 receptor 83 |
| chr3_-_103460529 | 4.87 |
ENSRNOT00000047274
|
LOC100909573
|
olfactory receptor 4F6-like |
| chr1_-_66212418 | 4.77 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr2_+_227080924 | 4.74 |
ENSRNOT00000029871
|
Fabp2
|
fatty acid binding protein 2 |
| chr5_-_58484900 | 4.73 |
ENSRNOT00000012386
|
Fam214b
|
family with sequence similarity 214, member B |
| chr1_+_62964901 | 4.61 |
ENSRNOT00000075013
|
LOC100365824
|
vomeronasal 2 receptor, 22-like |
| chrX_-_1704033 | 4.49 |
ENSRNOT00000051956
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr1_-_162713610 | 4.38 |
ENSRNOT00000018091
|
Aqp11
|
aquaporin 11 |
| chr19_+_15081590 | 4.31 |
ENSRNOT00000024187
|
Ces1f
|
carboxylesterase 1F |
| chr3_+_75465399 | 4.12 |
ENSRNOT00000057479
|
Olr562
|
olfactory receptor 562 |
| chrX_+_110014252 | 4.06 |
ENSRNOT00000093611
|
Nrk
|
Nik related kinase |
| chr7_-_98804499 | 4.06 |
ENSRNOT00000070909
|
Tatdn1
|
TatD DNase domain containing 1 |
| chr1_+_72230038 | 3.95 |
ENSRNOT00000058997
|
Vom1r38
|
vomeronasal 1 receptor 38 |
| chr3_-_77661863 | 3.91 |
ENSRNOT00000080807
|
Olr668
|
olfactory receptor 668 |
| chr3_-_148493225 | 3.81 |
ENSRNOT00000012141
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr9_+_7643533 | 3.79 |
ENSRNOT00000074923
|
Vom2r76
|
vomeronasal 2 receptor, 76 |
| chr18_+_61377051 | 3.77 |
ENSRNOT00000066659
|
Oacyl
|
O-acyltransferase like |
| chr3_+_78876609 | 3.73 |
ENSRNOT00000049936
|
Olr726
|
olfactory receptor 726 |
| chr9_+_93086012 | 3.57 |
ENSRNOT00000089470
|
Psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr3_-_103217528 | 3.52 |
ENSRNOT00000074456
|
Olr784
|
olfactory receptor 784 |
| chr7_-_57679795 | 3.46 |
ENSRNOT00000007461
|
Trhde
|
thyrotropin-releasing hormone degrading enzyme |
| chr1_-_172076138 | 3.44 |
ENSRNOT00000051241
|
Olr239
|
olfactory receptor 239 |
| chrX_+_83926513 | 3.36 |
ENSRNOT00000035274
|
RGD1561958
|
similar to RIKEN cDNA 2010106E10 |
| chr7_-_13425311 | 3.27 |
ENSRNOT00000060455
|
Olr1076
|
olfactory receptor 1076 |
| chr1_-_172215730 | 3.14 |
ENSRNOT00000055175
|
Olr244
|
olfactory receptor 244 |
| chr20_-_5927070 | 2.98 |
ENSRNOT00000059264
|
Slc26a8
|
solute carrier family 26 member 8 |
| chr4_+_118160147 | 2.87 |
ENSRNOT00000022014
|
Fam136a
|
family with sequence similarity 136, member A |
| chr9_+_84410972 | 2.86 |
ENSRNOT00000019724
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr3_+_153398130 | 2.85 |
ENSRNOT00000068135
|
Rpn2
|
ribophorin II |
| chr12_+_4248808 | 2.85 |
ENSRNOT00000042410
|
AABR07035089.1
|
|
| chr1_+_169616178 | 2.82 |
ENSRNOT00000023170
|
Olr157
|
olfactory receptor 157 |
| chrX_-_915953 | 2.69 |
ENSRNOT00000075264
|
Spaca5
|
sperm acrosome associated 5 |
| chr10_-_44082313 | 2.58 |
ENSRNOT00000051126
|
Olr1421
|
olfactory receptor 1421 |
| chr1_+_169919411 | 2.51 |
ENSRNOT00000050034
|
Olr186
|
olfactory receptor 186 |
| chr5_-_104920906 | 2.41 |
ENSRNOT00000071318
|
Fam154a
|
family with sequence similarity 154, member A |
| chr2_+_196608496 | 2.39 |
ENSRNOT00000091681
|
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr11_-_33760100 | 2.36 |
ENSRNOT00000065810
|
RGD1562683
|
RGD1562683 |
| chrX_-_45284341 | 2.35 |
ENSRNOT00000045436
|
Obp1f
|
odorant binding protein I f |
| chr7_-_143738237 | 2.34 |
ENSRNOT00000055320
|
Spryd3
|
SPRY domain containing 3 |
| chr3_-_78725950 | 2.33 |
ENSRNOT00000008690
|
Olr721
|
olfactory receptor 721 |
| chr4_+_87312766 | 2.26 |
ENSRNOT00000052126
|
Vom1r71
|
vomeronasal 1 receptor 71 |
| chr7_-_69104950 | 2.26 |
ENSRNOT00000049063
|
LOC500846
|
hypothetical protein LOC500846 |
| chr2_-_127781003 | 2.25 |
ENSRNOT00000050764
|
Mfsd8
|
major facilitator superfamily domain containing 8 |
| chr1_-_168725656 | 2.20 |
ENSRNOT00000040224
|
Olr115
|
olfactory receptor 115 |
| chr1_-_168819946 | 2.18 |
ENSRNOT00000048499
|
Olr121
|
olfactory receptor 121 |
| chr1_+_66663965 | 2.17 |
ENSRNOT00000040738
|
LOC108348135
|
vomeronasal type-1 receptor 2-like |
| chr2_-_100249811 | 2.16 |
ENSRNOT00000086760
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr3_-_102432479 | 2.15 |
ENSRNOT00000047270
|
Olr747
|
olfactory receptor 747 |
| chr4_-_117575154 | 2.09 |
ENSRNOT00000075813
|
LOC102556148
|
probable N-acetyltransferase CML2-like |
| chr11_+_83986230 | 2.05 |
ENSRNOT00000002331
ENSRNOT00000073703 |
Alg3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr3_+_79729739 | 2.05 |
ENSRNOT00000084833
|
Kbtbd4
|
kelch repeat and BTB domain containing 4 |
| chr1_-_99135977 | 2.05 |
ENSRNOT00000056515
|
Vom2r38
|
vomeronasal 2 receptor, 38 |
| chr1_-_88558387 | 2.02 |
ENSRNOT00000074174
|
AABR07002896.1
|
|
| chr13_-_98529040 | 2.01 |
ENSRNOT00000091715
|
Psen2
|
presenilin 2 |
| chr14_+_107178838 | 2.01 |
ENSRNOT00000091331
|
AABR07016619.1
|
|
| chr3_-_78942535 | 1.98 |
ENSRNOT00000008924
|
Olr731
|
olfactory receptor 731 |
| chr1_+_150492239 | 1.94 |
ENSRNOT00000047771
ENSRNOT00000048404 |
AABR07004684.1
|
|
| chr8_-_58253688 | 1.94 |
ENSRNOT00000010956
|
Cul5
|
cullin 5 |
| chr13_-_82129989 | 1.90 |
ENSRNOT00000078963
|
AC124874.1
|
|
| chr4_-_165774126 | 1.89 |
ENSRNOT00000007453
|
Tas2r107
|
taste receptor, type 2, member 107 |
| chr10_+_7279920 | 1.84 |
ENSRNOT00000003749
|
Tmem114
|
transmembrane protein 114 |
| chr4_-_97567844 | 1.83 |
ENSRNOT00000042169
|
Vom1r89
|
vomeronasal 1 receptor 89 |
| chr8_+_52751854 | 1.79 |
ENSRNOT00000072518
|
Nxpe1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr2_-_210874304 | 1.77 |
ENSRNOT00000088657
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr8_+_4405227 | 1.75 |
ENSRNOT00000040595
|
LOC100910798
|
vomeronasal type-2 receptor 26-like |
| chr7_-_4183525 | 1.74 |
ENSRNOT00000047673
|
Olr889
|
olfactory receptor 889 |
| chr18_-_36322320 | 1.74 |
ENSRNOT00000060260
|
Grxcr2
|
glutaredoxin and cysteine rich domain containing 2 |
| chr11_+_45462345 | 1.74 |
ENSRNOT00000029420
|
Nit2
|
nitrilase family, member 2 |
| chr1_+_116869957 | 1.72 |
ENSRNOT00000075203
|
Vom2r25
|
vomeronasal 2 receptor, 25 |
| chr2_+_4820016 | 1.69 |
ENSRNOT00000047222
ENSRNOT00000093226 |
Olr205
|
olfactory receptor 205 |
| chr17_-_44948432 | 1.65 |
ENSRNOT00000092195
|
LOC103690297
|
putative olfactory receptor 2W6 |
| chr1_+_150130084 | 1.65 |
ENSRNOT00000041243
|
Olr25
|
olfactory receptor 25 |
| chr11_-_45616429 | 1.63 |
ENSRNOT00000046587
|
Olr1533
|
olfactory receptor 1533 |
| chr8_+_41987849 | 1.57 |
ENSRNOT00000073800
|
LOC100911999
|
olfactory receptor 143-like |
| chr2_-_168260536 | 1.55 |
ENSRNOT00000072551
|
Vom1r55
|
vomeronasal 1 receptor 55 |
| chr2_+_168228099 | 1.53 |
ENSRNOT00000074681
|
Vom1r56
|
vomeronasal 1 receptor 56 |
| chr7_-_11754508 | 1.53 |
ENSRNOT00000026341
|
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr5_+_153845094 | 1.49 |
ENSRNOT00000041600
|
Stpg1
|
sperm-tail PG-rich repeat containing 1 |
| chr3_+_73988945 | 1.44 |
ENSRNOT00000050404
|
Olr515
|
olfactory receptor 515 |
| chr10_+_61160125 | 1.44 |
ENSRNOT00000074715
|
Olr1519
|
olfactory receptor 1519 |
| chr4_-_123161985 | 1.41 |
ENSRNOT00000011490
|
Xpc
|
XPC complex subunit, DNA damage recognition and repair factor |
| chr3_-_158985814 | 1.35 |
ENSRNOT00000071070
|
LOC100909423
|
olfactory receptor 150-like |
| chr3_+_102630019 | 1.33 |
ENSRNOT00000041799
|
Olr757
|
olfactory receptor 757 |
| chr8_-_19386384 | 1.29 |
ENSRNOT00000051572
|
Olr1145
|
olfactory receptor 1145 |
| chr3_+_76052230 | 1.28 |
ENSRNOT00000039963
|
LOC100911380
|
olfactory receptor 5W2-like |
| chr4_-_165828814 | 1.27 |
ENSRNOT00000007481
|
Tas2r105
|
taste receptor, type 2, member 105 |
| chr11_+_43521054 | 1.17 |
ENSRNOT00000041679
|
Olr1551
|
olfactory receptor 1551 |
| chr1_+_61132818 | 1.16 |
ENSRNOT00000041434
|
Vom1r20
|
vomeronasal 1 receptor 20 |
| chr1_+_142951094 | 1.13 |
ENSRNOT00000077441
|
Slc28a1
|
solute carrier family 28 member 1 |
| chr1_-_149529350 | 1.11 |
ENSRNOT00000052226
|
Vom2r43
|
vomeronasal 2 receptor, 43 |
| chr13_+_52147555 | 1.09 |
ENSRNOT00000084766
|
Lmod1
|
leiomodin 1 |
| chr10_-_91117889 | 1.07 |
ENSRNOT00000031498
|
Dcakd
|
dephospho-CoA kinase domain containing |
| chr16_-_52127591 | 1.05 |
ENSRNOT00000033652
|
Triml2
|
tripartite motif family-like 2 |
| chr10_+_60728160 | 1.05 |
ENSRNOT00000041085
|
Olr1501
|
olfactory receptor 1501 |
| chr12_+_52356832 | 0.98 |
ENSRNOT00000050252
|
Fbrsl1
|
fibrosin-like 1 |
| chr15_+_61879184 | 0.96 |
ENSRNOT00000042606
|
Sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr3_-_79051103 | 0.95 |
ENSRNOT00000088256
|
Olr737
|
olfactory receptor 737 |
| chrX_-_29825439 | 0.91 |
ENSRNOT00000048155
|
Gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr3_-_73561623 | 0.91 |
ENSRNOT00000052306
|
Olr486
|
olfactory receptor 486 |
| chr9_-_80295446 | 0.89 |
ENSRNOT00000023769
|
Tnp1
|
transition protein 1 |
| chr1_-_172465446 | 0.89 |
ENSRNOT00000088386
|
LOC103690410
|
olfactory receptor 484-like |
| chr8_-_21224737 | 0.86 |
ENSRNOT00000067321
|
Or7e24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr4_-_1752548 | 0.85 |
ENSRNOT00000071677
|
LOC100912415
|
olfactory receptor 150-like |
| chr1_-_220096319 | 0.83 |
ENSRNOT00000091787
ENSRNOT00000073983 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr17_+_25228437 | 0.83 |
ENSRNOT00000072904
|
AABR07027342.1
|
|
| chr3_+_78748001 | 0.79 |
ENSRNOT00000008715
|
Olr722
|
olfactory receptor 722 |
| chr6_+_44225233 | 0.76 |
ENSRNOT00000066593
|
Kidins220
|
kinase D-interacting substrate 220 |
| chr3_-_78193481 | 0.73 |
ENSRNOT00000008419
|
Olr696
|
olfactory receptor 696 |
| chr3_-_74014795 | 0.60 |
ENSRNOT00000073426
|
Olr516
|
olfactory receptor 516 |
| chr3_+_139894331 | 0.60 |
ENSRNOT00000064695
|
Rin2
|
Ras and Rab interactor 2 |
| chr16_-_75120417 | 0.59 |
ENSRNOT00000044094
|
Defb12
|
defensin beta 12 |
| chr8_-_23282989 | 0.58 |
ENSRNOT00000019204
|
Zfp717
|
zinc finger protein 717 |
| chr1_-_88558696 | 0.58 |
ENSRNOT00000038642
|
AABR07002896.1
|
|
| chr7_+_16958034 | 0.57 |
ENSRNOT00000034654
|
Vom2r54
|
vomeronasal 2 receptor, 54 |
| chr16_-_10603850 | 0.55 |
ENSRNOT00000087737
|
Fam35a
|
family with sequence similarity 35, member A |
| chr9_+_24170858 | 0.53 |
ENSRNOT00000081915
|
AC130064.1
|
|
| chr4_+_1513759 | 0.53 |
ENSRNOT00000073972
|
Olr1239
|
olfactory receptor 1239 |
| chr18_-_19275273 | 0.52 |
ENSRNOT00000041707
|
LOC102547344
|
lateral signaling target protein 2 homolog |
| chr10_+_73868943 | 0.51 |
ENSRNOT00000081012
|
Tubd1
|
tubulin, delta 1 |
| chr1_+_230450529 | 0.50 |
ENSRNOT00000045383
|
Olr371
|
olfactory receptor 371 |
| chr3_-_77618518 | 0.42 |
ENSRNOT00000044273
|
Olr666
|
olfactory receptor 666 |
| chr13_-_42885440 | 0.42 |
ENSRNOT00000038020
|
Nckap5
|
NCK-associated protein 5 |
| chr1_-_168587241 | 0.41 |
ENSRNOT00000021254
|
Olr104
|
olfactory receptor 104 |
| chr10_-_46172166 | 0.40 |
ENSRNOT00000091052
ENSRNOT00000004412 |
Flcn
|
folliculin |
| chr1_+_65409829 | 0.37 |
ENSRNOT00000046796
|
Vom2r19
|
vomeronasal 2 receptor, 19 |
| chr8_-_121973125 | 0.35 |
ENSRNOT00000012114
|
Pdcd6ip
|
programmed cell death 6 interacting protein |
| chr20_+_1861272 | 0.35 |
ENSRNOT00000044570
|
Olr1744
|
olfactory receptor 1744 |
| chr3_+_75771589 | 0.31 |
ENSRNOT00000007665
|
Olr598
|
olfactory receptor 598 |
| chr7_-_130151880 | 0.24 |
ENSRNOT00000088623
|
Plxnb2
|
plexin B2 |
| chr5_+_146656049 | 0.20 |
ENSRNOT00000036029
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr16_+_68633720 | 0.11 |
ENSRNOT00000081838
|
LOC100911229
|
sperm motility kinase-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Aire
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.4 | 62.3 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) |
| 8.3 | 24.9 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-12(GO:0071349) cellular response to interleukin-15(GO:0071350) |
| 5.5 | 16.4 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 3.5 | 24.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 3.4 | 27.2 | GO:0033590 | response to cobalamin(GO:0033590) |
| 3.0 | 9.1 | GO:0006742 | NADP catabolic process(GO:0006742) NAD catabolic process(GO:0019677) |
| 2.9 | 8.8 | GO:0015820 | leucine transport(GO:0015820) |
| 2.8 | 28.3 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 2.8 | 8.3 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 2.1 | 10.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.8 | 12.6 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 1.4 | 5.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 1.1 | 9.2 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 1.0 | 5.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 1.0 | 2.9 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.9 | 12.7 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.9 | 4.4 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.7 | 15.8 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.7 | 4.1 | GO:0060721 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.6 | 14.8 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.6 | 37.6 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.6 | 9.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.6 | 15.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.5 | 7.5 | GO:0036065 | fucosylation(GO:0036065) |
| 0.5 | 6.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.5 | 1.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.5 | 1.4 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.5 | 6.9 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.4 | 6.6 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.4 | 31.5 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.4 | 8.1 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.3 | 11.0 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.3 | 11.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.3 | 5.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 1.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 1.7 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.3 | 1.1 | GO:1904823 | purine nucleobase transmembrane transport(GO:1904823) |
| 0.2 | 12.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.2 | 1.0 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.2 | 6.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 1.9 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 14.7 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 2.0 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 2.4 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.2 | 0.4 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.2 | 14.9 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.2 | 3.0 | GO:0019532 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.2 | 9.1 | GO:0007588 | excretion(GO:0007588) |
| 0.2 | 4.1 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 11.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 28.8 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 8.6 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 9.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 1.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 5.7 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.4 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 4.7 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 0.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 3.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 3.2 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.1 | 11.7 | GO:0008037 | cell recognition(GO:0008037) |
| 0.1 | 7.6 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 2.1 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 90.1 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 1.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.8 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 8.5 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.1 | 0.8 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 2.2 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 2.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 7.8 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 3.8 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 8.0 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 2.1 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 1.7 | GO:0007605 | sensory perception of sound(GO:0007605) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:0090534 | calcium ion-transporting ATPase complex(GO:0090534) |
| 2.7 | 16.4 | GO:0032280 | symmetric synapse(GO:0032280) |
| 2.3 | 27.2 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 2.1 | 24.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.5 | 24.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.4 | 28.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.3 | 10.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.3 | 17.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 1.2 | 14.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 1.2 | 15.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 1.2 | 6.9 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 1.0 | 6.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.9 | 61.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.9 | 14.9 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.5 | 5.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.5 | 5.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.5 | 11.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.5 | 3.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.5 | 4.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.5 | 1.4 | GO:0071942 | XPC complex(GO:0071942) |
| 0.5 | 12.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 8.0 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.3 | 1.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 7.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 12.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 5.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 2.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 3.6 | GO:0022624 | proteasome accessory complex(GO:0022624) |
| 0.1 | 75.0 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 25.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 8.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 7.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 9.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 7.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 7.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 93.5 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 0.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 4.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 7.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 2.0 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.0 | 19.4 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 31.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 4.4 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 6.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.4 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 1.1 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.8 | 62.3 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 3.4 | 27.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 3.0 | 14.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 2.8 | 8.3 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 2.7 | 16.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 2.3 | 9.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 2.3 | 61.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 2.2 | 11.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 1.9 | 24.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 1.9 | 5.7 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) |
| 1.8 | 14.7 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.8 | 12.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 1.7 | 5.1 | GO:0032142 | dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 1.7 | 14.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.5 | 9.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 1.3 | 7.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 1.1 | 24.9 | GO:0005521 | phosphatidylinositol phospholipase C activity(GO:0004435) lamin binding(GO:0005521) |
| 0.8 | 10.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.8 | 24.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.8 | 4.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.7 | 11.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.7 | 2.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.7 | 8.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.6 | 13.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.5 | 1.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.5 | 9.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.5 | 2.9 | GO:0004144 | 2-acylglycerol O-acyltransferase activity(GO:0003846) diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.5 | 5.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.4 | 10.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.4 | 1.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 6.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.4 | 1.1 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.4 | 4.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.4 | 6.9 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 7.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 8.8 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.3 | 4.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 29.6 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.3 | 16.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 1.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.2 | 8.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 2.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 2.8 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 3.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 15.2 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.2 | 2.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.2 | 9.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 0.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 1.9 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 0.8 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 3.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 14.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 11.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 1.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 8.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 2.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 10.1 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 6.5 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 1.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 5.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 7.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 5.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 4.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 60.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 4.1 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 2.2 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 4.3 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.7 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 10.5 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 4.1 | GO:0016747 | transferase activity, transferring acyl groups other than amino-acyl groups(GO:0016747) |
| 0.0 | 2.3 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 24.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.5 | 5.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 12.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.4 | 10.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.4 | 14.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 6.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 5.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 8.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 6.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 7.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 57.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 2.5 | 24.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.8 | 14.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.7 | 10.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.7 | 11.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.6 | 14.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.5 | 8.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.4 | 14.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 11.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 6.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 11.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 5.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 1.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 10.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 6.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 2.0 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 8.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 4.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 1.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 5.5 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 2.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 2.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 9.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 2.8 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 8.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 1.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |