Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Zkscan3
Z-value: 0.33
Transcription factors associated with Zkscan3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zkscan3
|
ENSRNOG00000055000 | zinc finger with KRAB and SCAN domains 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zkscan3 | rn6_v1_chr17_+_45247776_45247794 | -0.44 | 4.6e-01 | Click! |
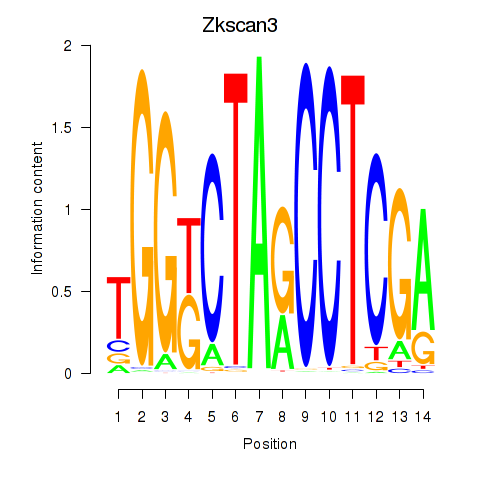
Activity profile of Zkscan3 motif
Sorted Z-values of Zkscan3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_47698947 | 0.18 |
ENSRNOT00000001586
|
Trpv4
|
transient receptor potential cation channel, subfamily V, member 4 |
| chr8_+_116776494 | 0.18 |
ENSRNOT00000050702
|
Cdhr4
|
cadherin-related family member 4 |
| chr19_-_43911057 | 0.12 |
ENSRNOT00000026017
|
Ctrb1
|
chymotrypsinogen B1 |
| chr7_-_70452675 | 0.11 |
ENSRNOT00000090498
|
AC114111.1
|
|
| chr14_+_10692764 | 0.08 |
ENSRNOT00000003012
|
LOC100910270
|
uncharacterized LOC100910270 |
| chr1_+_80426918 | 0.07 |
ENSRNOT00000067272
|
Nkpd1
|
NTPase, KAP family P-loop domain containing 1 |
| chr7_-_12432130 | 0.07 |
ENSRNOT00000077301
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr16_+_19792838 | 0.07 |
ENSRNOT00000022938
|
Babam1
|
BRISC and BRCA1 A complex member 1 |
| chr13_+_49074644 | 0.06 |
ENSRNOT00000000041
|
Klhdc8a
|
kelch domain containing 8A |
| chr2_+_197682000 | 0.06 |
ENSRNOT00000066821
|
Hormad1
|
HORMA domain containing 1 |
| chr5_+_157434481 | 0.06 |
ENSRNOT00000088556
|
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr1_-_221917901 | 0.06 |
ENSRNOT00000092270
|
Slc22a12
|
solute carrier family 22 member 12 |
| chr7_-_140770647 | 0.05 |
ENSRNOT00000081784
|
C1ql4
|
complement C1q like 4 |
| chr3_+_11679530 | 0.05 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr20_+_13156241 | 0.05 |
ENSRNOT00000050531
|
Prmt2
|
protein arginine methyltransferase 2 |
| chr5_+_157642751 | 0.05 |
ENSRNOT00000083577
ENSRNOT00000010308 |
Capzb
|
capping actin protein of muscle Z-line beta subunit |
| chr8_+_116096458 | 0.04 |
ENSRNOT00000021188
|
RGD1307461
|
similar to RIKEN cDNA 6430571L13 gene; similar to g20 protein |
| chr1_+_78671121 | 0.04 |
ENSRNOT00000021310
|
Ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr10_-_13051154 | 0.04 |
ENSRNOT00000033457
|
AABR07029202.1
|
|
| chr13_+_99214389 | 0.04 |
ENSRNOT00000004354
|
Lefty1
|
left right determination factor 1 |
| chr12_-_18540166 | 0.04 |
ENSRNOT00000001792
|
Il3ra
|
interleukin 3 receptor subunit alpha |
| chr10_-_62254287 | 0.04 |
ENSRNOT00000004313
|
Serpinf1
|
serpin family F member 1 |
| chr3_-_10153554 | 0.04 |
ENSRNOT00000093246
|
Exosc2
|
exosome component 2 |
| chr9_-_10338216 | 0.03 |
ENSRNOT00000070862
|
Ndufa11
|
NADH:ubiquinone oxidoreductase subunit A11 |
| chr9_-_71852113 | 0.03 |
ENSRNOT00000083263
ENSRNOT00000072983 |
AABR07067896.1
|
|
| chr8_+_71719563 | 0.03 |
ENSRNOT00000022828
|
Ppib
|
peptidylprolyl isomerase B |
| chr15_-_58711872 | 0.03 |
ENSRNOT00000058204
|
Serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr4_-_148845267 | 0.03 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr6_-_117972898 | 0.03 |
ENSRNOT00000032968
|
AABR07065265.1
|
|
| chr10_+_105861743 | 0.03 |
ENSRNOT00000064410
|
Mgat5b
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr13_-_52136127 | 0.03 |
ENSRNOT00000009398
|
Timm17a
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr7_+_11301957 | 0.02 |
ENSRNOT00000027847
|
Tjp3
|
tight junction protein 3 |
| chr13_+_48427038 | 0.02 |
ENSRNOT00000009241
|
Ctse
|
cathepsin E |
| chr10_+_106065712 | 0.02 |
ENSRNOT00000003690
|
Sec14l1
|
SEC14-like lipid binding 1 |
| chr1_-_226924244 | 0.02 |
ENSRNOT00000028971
|
Tmem132a
|
transmembrane protein 132A |
| chr1_-_242373764 | 0.02 |
ENSRNOT00000076544
|
Fam122a
|
family with sequence similarity 122A |
| chr7_-_119200623 | 0.02 |
ENSRNOT00000029753
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr3_-_23297774 | 0.02 |
ENSRNOT00000019162
|
Rpl35
|
ribosomal protein L35 |
| chr9_+_49479023 | 0.02 |
ENSRNOT00000050922
ENSRNOT00000077111 |
Pou3f3
|
POU class 3 homeobox 3 |
| chr17_+_47662532 | 0.02 |
ENSRNOT00000081314
|
Stard3nl
|
STARD3 N-terminal like |
| chr9_-_82461903 | 0.02 |
ENSRNOT00000026654
|
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr10_+_14216155 | 0.02 |
ENSRNOT00000020192
|
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr10_-_14215957 | 0.01 |
ENSRNOT00000019767
|
Fahd1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr8_-_59607275 | 0.01 |
ENSRNOT00000019307
|
Chrna3
|
cholinergic receptor nicotinic alpha 3 subunit |
| chr14_-_89179507 | 0.01 |
ENSRNOT00000006498
|
Pkd1l1
|
polycystin 1 like 1, transient receptor potential channel interacting |
| chr1_+_277190964 | 0.01 |
ENSRNOT00000080511
|
Casp7
|
caspase 7 |
| chr5_+_24146171 | 0.01 |
ENSRNOT00000037547
|
MGC94199
|
similar to RIKEN cDNA 2610301B20; EST AI428449 |
| chr10_-_34961349 | 0.01 |
ENSRNOT00000004885
|
LOC103689931
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr19_-_57649827 | 0.01 |
ENSRNOT00000026757
|
Exoc8
|
exocyst complex component 8 |
| chr10_-_37055282 | 0.01 |
ENSRNOT00000086193
ENSRNOT00000065584 |
Hnrnpab
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr4_-_161907767 | 0.01 |
ENSRNOT00000009557
|
A2ml1
|
alpha-2-macroglobulin-like 1 |
| chr18_+_28409955 | 0.01 |
ENSRNOT00000026989
|
Paip2
|
poly(A) binding protein interacting protein 2 |
| chr2_-_59181863 | 0.01 |
ENSRNOT00000079636
|
Spef2
|
sperm flagellar 2 |
| chr12_-_47095438 | 0.01 |
ENSRNOT00000001546
|
Coq5
|
coenzyme Q5, methyltransferase |
| chr5_+_23358841 | 0.01 |
ENSRNOT00000070963
|
MGC94199
|
similar to RIKEN cDNA 2610301B20; EST AI428449 |
| chr15_-_45550285 | 0.01 |
ENSRNOT00000012948
|
Gucy1b2
|
guanylate cyclase 1 soluble subunit beta 2 |
| chr1_+_99532568 | 0.00 |
ENSRNOT00000077073
|
Zfp819
|
zinc finger protein 819 |
| chrX_-_139229702 | 0.00 |
ENSRNOT00000042507
|
AABR07041771.1
|
|
| chr1_-_174620064 | 0.00 |
ENSRNOT00000016224
|
Tmem41b
|
transmembrane protein 41B |
| chr7_+_70452579 | 0.00 |
ENSRNOT00000046099
|
B4galnt1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr8_+_79722334 | 0.00 |
ENSRNOT00000083957
|
Rab27a
|
RAB27A, member RAS oncogene family |
| chr11_-_69355854 | 0.00 |
ENSRNOT00000002975
|
Ropn1
|
rhophilin associated tail protein 1 |
| chr19_+_49462129 | 0.00 |
ENSRNOT00000015158
|
Cenpn
|
centromere protein N |
| chr7_+_64672722 | 0.00 |
ENSRNOT00000064448
ENSRNOT00000005539 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr2_-_59084059 | 0.00 |
ENSRNOT00000086323
ENSRNOT00000088701 |
Spef2
|
sperm flagellar 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zkscan3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.1 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.0 | 0.0 | GO:0071031 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.0 | 0.0 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.0 | 0.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |