Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Zic4
Z-value: 0.54
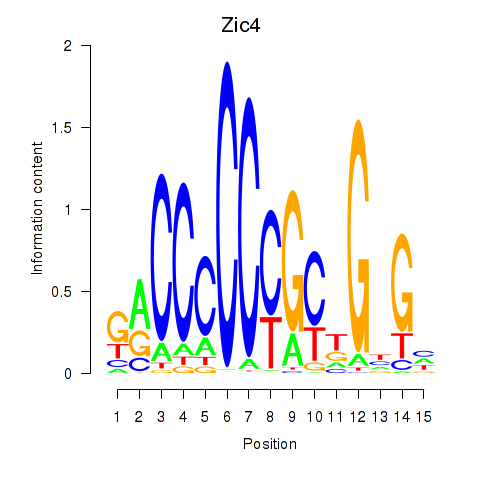
Transcription factors associated with Zic4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zic4
|
ENSRNOG00000014871 | Zic family member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic4 | rn6_v1_chr8_+_98745310_98745310 | 0.79 | 1.1e-01 | Click! |
Activity profile of Zic4 motif
Sorted Z-values of Zic4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_20607507 | 0.50 |
ENSRNOT00000000011
|
Cbln1
|
cerebellin 1 precursor |
| chr17_-_9695292 | 0.47 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr10_-_40953651 | 0.42 |
ENSRNOT00000063889
|
Glra1
|
glycine receptor, alpha 1 |
| chr6_+_33885495 | 0.42 |
ENSRNOT00000086633
|
Sdc1
|
syndecan 1 |
| chr10_-_40953467 | 0.28 |
ENSRNOT00000092189
|
Glra1
|
glycine receptor, alpha 1 |
| chr1_-_67390141 | 0.27 |
ENSRNOT00000025808
|
Sbk1
|
SH3 domain binding kinase 1 |
| chr16_-_20890949 | 0.26 |
ENSRNOT00000081977
|
Homer3
|
homer scaffolding protein 3 |
| chr1_-_209641123 | 0.26 |
ENSRNOT00000021702
|
Ebf3
|
early B-cell factor 3 |
| chr8_-_68038533 | 0.26 |
ENSRNOT00000018756
|
Skor1
|
SKI family transcriptional corepressor 1 |
| chr3_+_147585947 | 0.24 |
ENSRNOT00000006833
|
Scrt2
|
scratch family transcriptional repressor 2 |
| chr3_-_2803574 | 0.23 |
ENSRNOT00000040995
|
RGD1560470
|
similar to Gene model 996 |
| chr9_+_82120059 | 0.23 |
ENSRNOT00000057368
|
Cdk5r2
|
cyclin-dependent kinase 5 regulatory subunit 2 |
| chr9_-_79545024 | 0.20 |
ENSRNOT00000021152
|
Mreg
|
melanoregulin |
| chr1_+_197659187 | 0.19 |
ENSRNOT00000082228
|
LOC103690016
|
serine/threonine-protein kinase SBK1 |
| chr1_+_266953139 | 0.18 |
ENSRNOT00000054696
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr9_-_119653255 | 0.17 |
ENSRNOT00000056272
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr5_+_135962911 | 0.15 |
ENSRNOT00000087353
|
Ptch2
|
patched 2 |
| chr13_+_71107465 | 0.12 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr1_+_185356975 | 0.12 |
ENSRNOT00000086681
|
Plekha7
|
pleckstrin homology domain containing A7 |
| chr6_-_126582034 | 0.10 |
ENSRNOT00000010656
ENSRNOT00000080829 |
Itpk1
|
inositol-tetrakisphosphate 1-kinase |
| chr9_+_94980409 | 0.10 |
ENSRNOT00000035338
|
Dgkd
|
diacylglycerol kinase, delta |
| chr11_+_71872830 | 0.10 |
ENSRNOT00000050364
|
Nrros
|
negative regulator of reactive oxygen species |
| chr3_-_148428288 | 0.08 |
ENSRNOT00000072663
|
Dusp15
|
dual specificity phosphatase 15 |
| chr4_-_82160240 | 0.08 |
ENSRNOT00000038775
ENSRNOT00000058985 |
Hoxa4
|
homeo box A4 |
| chr10_+_48240127 | 0.08 |
ENSRNOT00000080682
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr1_-_216663720 | 0.08 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr9_-_16519144 | 0.07 |
ENSRNOT00000072581
|
LOC681367
|
hypothetical protein LOC681367 |
| chr16_-_20686317 | 0.07 |
ENSRNOT00000060097
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr3_+_112916217 | 0.07 |
ENSRNOT00000030065
|
Tmem62
|
transmembrane protein 62 |
| chr1_-_78997869 | 0.07 |
ENSRNOT00000023490
|
Hif3a
|
hypoxia inducible factor 3, alpha subunit |
| chr13_-_51076165 | 0.07 |
ENSRNOT00000004602
|
Adora1
|
adenosine A1 receptor |
| chrX_-_157312028 | 0.06 |
ENSRNOT00000077979
|
Atp2b3
|
ATPase plasma membrane Ca2+ transporting 3 |
| chr14_-_84170301 | 0.06 |
ENSRNOT00000080413
|
Slc35e4
|
solute carrier family 35, member E4 |
| chr7_-_144960527 | 0.06 |
ENSRNOT00000086554
|
Zfp385a
|
zinc finger protein 385A |
| chr10_+_106812739 | 0.06 |
ENSRNOT00000074225
|
Syngr2
|
synaptogyrin 2 |
| chr1_+_226435979 | 0.06 |
ENSRNOT00000048704
ENSRNOT00000036232 ENSRNOT00000035576 ENSRNOT00000036180 ENSRNOT00000036168 ENSRNOT00000047964 ENSRNOT00000036283 ENSRNOT00000007429 |
Syt7
|
synaptotagmin 7 |
| chr7_+_116943057 | 0.06 |
ENSRNOT00000056527
|
Tigd5
|
tigger transposable element derived 5 |
| chr4_-_77747070 | 0.06 |
ENSRNOT00000009247
|
Zfp746
|
zinc finger protein 746 |
| chr20_-_3822754 | 0.05 |
ENSRNOT00000000541
ENSRNOT00000077357 |
Slc39a7
|
solute carrier family 39 member 7 |
| chr2_+_187512164 | 0.05 |
ENSRNOT00000051394
|
Mef2d
|
myocyte enhancer factor 2D |
| chr3_+_110982553 | 0.05 |
ENSRNOT00000030754
|
LOC691418
|
hypothetical protein LOC691418 |
| chr2_+_95008477 | 0.05 |
ENSRNOT00000015327
|
Tpd52
|
tumor protein D52 |
| chr7_+_130474279 | 0.05 |
ENSRNOT00000092388
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr10_-_75270561 | 0.05 |
ENSRNOT00000012279
|
Dynll2
|
dynein light chain LC8-type 2 |
| chr10_+_48240330 | 0.05 |
ENSRNOT00000057798
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr7_+_130474508 | 0.05 |
ENSRNOT00000085191
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr1_-_204605552 | 0.05 |
ENSRNOT00000022904
|
Nkx1-2
|
NK1 homeobox 2 |
| chr1_-_212549477 | 0.05 |
ENSRNOT00000024765
|
Caly
|
calcyon neuron-specific vesicular protein |
| chr6_-_6842758 | 0.05 |
ENSRNOT00000006094
|
Kcng3
|
potassium voltage-gated channel modifier subfamily G member 3 |
| chr10_+_43224269 | 0.04 |
ENSRNOT00000003473
|
Sap30l
|
SAP30-like |
| chr19_-_43596801 | 0.04 |
ENSRNOT00000025625
|
Fa2h
|
fatty acid 2-hydroxylase |
| chr9_+_10956056 | 0.04 |
ENSRNOT00000073930
|
Plin5
|
perilipin 5 |
| chr8_-_36760742 | 0.04 |
ENSRNOT00000017307
|
Ddx25
|
DEAD-box helicase 25 |
| chr5_-_150323063 | 0.04 |
ENSRNOT00000014084
|
Oprd1
|
opioid receptor, delta 1 |
| chr10_-_91047177 | 0.04 |
ENSRNOT00000003986
|
C1ql1
|
complement C1q like 1 |
| chr16_+_59197367 | 0.04 |
ENSRNOT00000014924
|
Dlc1
|
DLC1 Rho GTPase activating protein |
| chr7_-_139318455 | 0.03 |
ENSRNOT00000092029
|
Hdac7
|
histone deacetylase 7 |
| chr3_+_175885894 | 0.03 |
ENSRNOT00000039780
|
Ntsr1
|
neurotensin receptor 1 |
| chr2_-_211831551 | 0.03 |
ENSRNOT00000039225
ENSRNOT00000090561 |
Fam102b
|
family with sequence similarity 102, member B |
| chr10_-_46145548 | 0.03 |
ENSRNOT00000033483
|
Pld6
|
phospholipase D family, member 6 |
| chr1_-_165967069 | 0.03 |
ENSRNOT00000089359
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr6_+_69950500 | 0.03 |
ENSRNOT00000049500
|
RGD1559629
|
similar to H+ ATP synthase |
| chr7_+_117963740 | 0.03 |
ENSRNOT00000075405
|
LOC108348189
|
COMM domain-containing protein 5 |
| chr10_-_105795958 | 0.03 |
ENSRNOT00000067739
|
Srsf2
|
serine and arginine rich splicing factor 2 |
| chr13_+_110920830 | 0.03 |
ENSRNOT00000077014
ENSRNOT00000076362 |
Kcnh1
|
potassium voltage-gated channel subfamily H member 1 |
| chr19_+_37600148 | 0.02 |
ENSRNOT00000023853
|
Ctcf
|
CCCTC-binding factor |
| chr8_+_117246376 | 0.02 |
ENSRNOT00000074493
|
Ccdc71
|
coiled-coil domain containing 71 |
| chr1_+_216293087 | 0.02 |
ENSRNOT00000027875
ENSRNOT00000087153 |
Kcnq1
|
potassium voltage-gated channel subfamily Q member 1 |
| chr1_-_199270627 | 0.02 |
ENSRNOT00000026063
|
Stx1b
|
syntaxin 1B |
| chr3_+_72540538 | 0.02 |
ENSRNOT00000012379
|
Aplnr
|
apelin receptor |
| chr10_+_80790168 | 0.02 |
ENSRNOT00000073315
ENSRNOT00000075163 |
Car10
|
carbonic anhydrase 10 |
| chr19_+_44164935 | 0.02 |
ENSRNOT00000048998
|
Gabarapl2
|
GABA type A receptor associated protein like 2 |
| chr2_+_231962517 | 0.02 |
ENSRNOT00000014570
|
Neurog2
|
neurogenin 2 |
| chr5_+_154112076 | 0.02 |
ENSRNOT00000044969
|
Myom3
|
myomesin 3 |
| chr16_+_59988603 | 0.02 |
ENSRNOT00000015176
|
Cldn23
|
claudin 23 |
| chr10_-_92008082 | 0.02 |
ENSRNOT00000006361
|
Nsf
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr18_+_3995921 | 0.02 |
ENSRNOT00000091424
|
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr16_+_17538855 | 0.02 |
ENSRNOT00000014772
|
Tspan14
|
tetraspanin 14 |
| chr1_-_65551043 | 0.02 |
ENSRNOT00000029996
|
Trim28
|
tripartite motif-containing 28 |
| chr3_+_3310954 | 0.02 |
ENSRNOT00000061773
|
Kcnt1
|
potassium sodium-activated channel subfamily T member 1 |
| chr5_+_159914931 | 0.01 |
ENSRNOT00000037202
|
Fam131c
|
family with sequence similarity 131, member C |
| chr7_-_75422268 | 0.01 |
ENSRNOT00000080218
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr4_+_49369296 | 0.01 |
ENSRNOT00000007822
|
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr19_+_39229754 | 0.01 |
ENSRNOT00000050612
|
Vps4a
|
vacuolar protein sorting 4 homolog A |
| chr1_+_219348721 | 0.01 |
ENSRNOT00000025084
|
Pitpnm1
|
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr7_+_144078496 | 0.01 |
ENSRNOT00000055302
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr1_-_80689171 | 0.01 |
ENSRNOT00000045574
|
Bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr14_+_86101277 | 0.01 |
ENSRNOT00000018846
|
Aebp1
|
AE binding protein 1 |
| chr8_+_50310405 | 0.01 |
ENSRNOT00000073507
|
Sik3
|
SIK family kinase 3 |
| chr1_+_154377447 | 0.01 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr3_-_148428494 | 0.01 |
ENSRNOT00000011350
|
Dusp15
|
dual specificity phosphatase 15 |
| chr5_-_28131133 | 0.01 |
ENSRNOT00000067331
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr2_+_95008311 | 0.01 |
ENSRNOT00000077270
|
Tpd52
|
tumor protein D52 |
| chr1_-_84491466 | 0.01 |
ENSRNOT00000034609
|
Map3k10
|
mitogen activated protein kinase kinase kinase 10 |
| chr11_+_75434197 | 0.01 |
ENSRNOT00000032569
|
Mb21d2
|
Mab-21 domain containing 2 |
| chr10_+_105498728 | 0.01 |
ENSRNOT00000032163
|
Sphk1
|
sphingosine kinase 1 |
| chr7_+_140788987 | 0.01 |
ENSRNOT00000086611
|
Kcnh3
|
potassium voltage-gated channel subfamily H member 3 |
| chr5_-_40237591 | 0.01 |
ENSRNOT00000011393
|
Fut9
|
fucosyltransferase 9 |
| chr12_+_25036605 | 0.01 |
ENSRNOT00000001996
ENSRNOT00000084427 |
Limk1
|
LIM domain kinase 1 |
| chr7_-_139254149 | 0.01 |
ENSRNOT00000078644
|
Rapgef3
|
Rap guanine nucleotide exchange factor 3 |
| chr15_-_33766438 | 0.01 |
ENSRNOT00000033977
|
Ap1g2
|
adaptor-related protein complex 1, gamma 2 subunit |
| chr7_+_116355911 | 0.01 |
ENSRNOT00000076182
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr4_+_175431904 | 0.01 |
ENSRNOT00000032843
|
Pde3a
|
phosphodiesterase 3A |
| chr7_+_54031316 | 0.00 |
ENSRNOT00000039096
|
Bbs10
|
Bardet-Biedl syndrome 10 |
| chr13_-_48284990 | 0.00 |
ENSRNOT00000086928
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr8_+_67295727 | 0.00 |
ENSRNOT00000020443
|
Anp32a
|
acidic nuclear phosphoprotein 32 family member A |
| chr10_+_6975244 | 0.00 |
ENSRNOT00000046232
|
Usp7
|
ubiquitin specific peptidase 7 |
| chr6_-_50786967 | 0.00 |
ENSRNOT00000009566
|
Cbll1
|
Cbl proto-oncogene like 1 |
| chr4_+_34238440 | 0.00 |
ENSRNOT00000010952
|
Mios
|
meiosis regulator for oocyte development |
| chr16_+_23317953 | 0.00 |
ENSRNOT00000075287
|
AABR07024972.1
|
|
| chr4_+_132137793 | 0.00 |
ENSRNOT00000014455
|
Gpr27
|
G protein-coupled receptor 27 |
| chr5_-_74191167 | 0.00 |
ENSRNOT00000088169
|
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr7_-_117786872 | 0.00 |
ENSRNOT00000030517
|
Lrrc24
|
leucine rich repeat containing 24 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zic4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.7 | GO:2000344 | negative regulation of transmission of nerve impulse(GO:0051970) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.2 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.5 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) negative regulation of long term synaptic depression(GO:1900453) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.0 | GO:0089718 | amino acid import across plasma membrane(GO:0089718) |
| 0.0 | 0.0 | GO:0007314 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0030977 | taurine binding(GO:0030977) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.0 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.1 | GO:0051766 | inositol trisphosphate kinase activity(GO:0051766) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |