Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
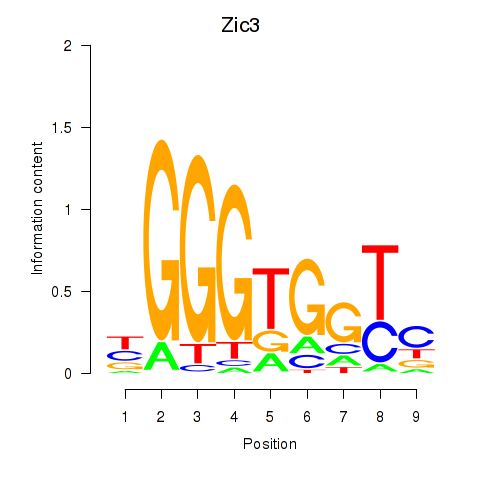
Results for Zic3
Z-value: 0.26
Transcription factors associated with Zic3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zic3
|
ENSRNOG00000000861 | Zic family member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic3 | rn6_v1_chrX_+_140878216_140878216 | -0.36 | 5.5e-01 | Click! |
Activity profile of Zic3 motif
Sorted Z-values of Zic3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_14348046 | 0.24 |
ENSRNOT00000018630
|
Cdhr1
|
cadherin-related family member 1 |
| chr8_+_44136496 | 0.09 |
ENSRNOT00000087022
|
Scn3b
|
sodium voltage-gated channel beta subunit 3 |
| chr3_+_80833272 | 0.08 |
ENSRNOT00000023583
|
Chrm4
|
cholinergic receptor, muscarinic 4 |
| chrX_+_76786466 | 0.08 |
ENSRNOT00000090665
|
Fgf16
|
fibroblast growth factor 16 |
| chr18_-_28454756 | 0.07 |
ENSRNOT00000040091
|
Spata24
|
spermatogenesis associated 24 |
| chr3_-_39596718 | 0.07 |
ENSRNOT00000006784
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr7_+_12006710 | 0.06 |
ENSRNOT00000045421
|
Klf16
|
Kruppel-like factor 16 |
| chr7_-_143967484 | 0.06 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr6_-_95502775 | 0.05 |
ENSRNOT00000074990
ENSRNOT00000034289 |
Dhrs7l1
|
dehydrogenase/reductase (SDR family) member 7-like 1 |
| chr4_+_115416580 | 0.05 |
ENSRNOT00000018303
|
Atp6v1b1
|
ATPase H+ transporting V1 subunit B1 |
| chr7_-_61798729 | 0.05 |
ENSRNOT00000010283
|
Dyrk2
|
dual specificity tyrosine phosphorylation regulated kinase 2 |
| chr2_+_216863428 | 0.05 |
ENSRNOT00000068413
|
Col11a1
|
collagen type XI alpha 1 chain |
| chr19_-_11144641 | 0.05 |
ENSRNOT00000081246
|
Slc12a3
|
solute carrier family 12 member 3 |
| chr10_+_59566223 | 0.05 |
ENSRNOT00000024068
ENSRNOT00000088245 |
P2rx1
|
purinergic receptor P2X 1 |
| chr10_+_3227160 | 0.05 |
ENSRNOT00000088075
|
Ntan1
|
N-terminal asparagine amidase |
| chr13_+_52887649 | 0.05 |
ENSRNOT00000048033
|
Ascl5
|
achaete-scute family bHLH transcription factor 5 |
| chr6_+_27975417 | 0.05 |
ENSRNOT00000077830
|
Dtnb
|
dystrobrevin, beta |
| chr16_+_22979444 | 0.05 |
ENSRNOT00000017822
|
Csgalnact1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr13_-_70626252 | 0.05 |
ENSRNOT00000036947
|
Lamc2
|
laminin subunit gamma 2 |
| chr7_-_143966863 | 0.05 |
ENSRNOT00000018828
|
Sp7
|
Sp7 transcription factor |
| chr17_+_43458553 | 0.04 |
ENSRNOT00000088939
|
Slc17a4
|
solute carrier family 17, member 4 |
| chr7_-_121311565 | 0.04 |
ENSRNOT00000092252
|
Rpl3
|
ribosomal protein L3 |
| chr8_-_53362006 | 0.04 |
ENSRNOT00000077145
|
LOC100360390
|
claudin 25-like |
| chr2_-_260596777 | 0.04 |
ENSRNOT00000009370
|
Lhx8
|
LIM homeobox 8 |
| chr1_-_88989552 | 0.04 |
ENSRNOT00000034001
|
Arhgap33
|
Rho GTPase activating protein 33 |
| chr9_-_55673704 | 0.04 |
ENSRNOT00000066231
ENSRNOT00000081677 |
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr9_-_15700235 | 0.04 |
ENSRNOT00000088713
ENSRNOT00000035907 |
Trerf1
|
transcriptional regulating factor 1 |
| chr10_-_90312386 | 0.04 |
ENSRNOT00000028445
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr10_-_110585376 | 0.04 |
ENSRNOT00000054917
|
Rab40b
|
Rab40b, member RAS oncogene family |
| chr7_+_41115154 | 0.04 |
ENSRNOT00000066174
|
Atp2b1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr19_+_60017746 | 0.04 |
ENSRNOT00000042623
|
Pard3
|
par-3 family cell polarity regulator |
| chr20_-_47306318 | 0.03 |
ENSRNOT00000075151
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr3_+_100787449 | 0.03 |
ENSRNOT00000078543
|
Bdnf
|
brain-derived neurotrophic factor |
| chr10_-_87541851 | 0.03 |
ENSRNOT00000089610
|
RGD1561684
|
similar to keratin associated protein 2-4 |
| chr2_+_199283909 | 0.03 |
ENSRNOT00000043194
|
Acp6
|
acid phosphatase 6, lysophosphatidic |
| chr16_+_19800463 | 0.03 |
ENSRNOT00000023065
|
Ankle1
|
ankyrin repeat and LEM domain containing 1 |
| chr10_-_13580821 | 0.03 |
ENSRNOT00000009735
|
Ntn3
|
netrin 3 |
| chr2_+_195582781 | 0.03 |
ENSRNOT00000066020
|
Them5
|
thioesterase superfamily member 5 |
| chr3_-_10196626 | 0.03 |
ENSRNOT00000012234
|
Prdm12
|
PR/SET domain 12 |
| chr20_-_4401610 | 0.03 |
ENSRNOT00000091468
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr1_-_176607466 | 0.03 |
ENSRNOT00000022984
|
Galnt18
|
polypeptide N-acetylgalactosaminyltransferase 18 |
| chr5_+_76182121 | 0.03 |
ENSRNOT00000020707
|
Gng10
|
G protein subunit gamma 10 |
| chr8_-_48774898 | 0.03 |
ENSRNOT00000016303
|
Upk2
|
uroplakin 2 |
| chr7_+_12764993 | 0.03 |
ENSRNOT00000017064
|
Grin3b
|
glutamate ionotropic receptor NMDA type subunit 3B |
| chr2_+_9683570 | 0.03 |
ENSRNOT00000074157
|
Cetn3
|
centrin 3 |
| chr8_-_129919120 | 0.03 |
ENSRNOT00000074423
|
Ulk4
|
unc-51 like kinase 4 |
| chr10_-_35005287 | 0.03 |
ENSRNOT00000032386
|
LOC103689949
|
nedd4 binding protein 3 |
| chr20_+_4576514 | 0.03 |
ENSRNOT00000090125
ENSRNOT00000047370 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chr12_+_48598647 | 0.03 |
ENSRNOT00000000889
|
Tmem119
|
transmembrane protein 119 |
| chr3_+_175885894 | 0.03 |
ENSRNOT00000039780
|
Ntsr1
|
neurotensin receptor 1 |
| chr13_+_52624878 | 0.03 |
ENSRNOT00000076054
ENSRNOT00000076299 |
Tnni1
|
troponin I1, slow skeletal type |
| chr5_-_173653905 | 0.03 |
ENSRNOT00000038556
|
Plekhn1
|
pleckstrin homology domain containing N1 |
| chr3_+_5624506 | 0.03 |
ENSRNOT00000036995
|
Adamtsl2
|
ADAMTS-like 2 |
| chr1_-_72461547 | 0.02 |
ENSRNOT00000022480
|
Ssc5d
|
scavenger receptor cysteine rich family member with 5 domains |
| chr20_-_4390436 | 0.02 |
ENSRNOT00000000497
ENSRNOT00000077655 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr3_+_111545007 | 0.02 |
ENSRNOT00000007247
|
Itpka
|
inositol-trisphosphate 3-kinase A |
| chr1_+_81230612 | 0.02 |
ENSRNOT00000026489
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr1_-_228753422 | 0.02 |
ENSRNOT00000028626
|
Dtx4
|
deltex E3 ubiquitin ligase 4 |
| chr10_+_56260514 | 0.02 |
ENSRNOT00000016177
|
Sox15
|
SRY box 15 |
| chr1_-_80744831 | 0.02 |
ENSRNOT00000025913
|
Bcl3
|
B-cell CLL/lymphoma 3 |
| chr5_+_153507093 | 0.02 |
ENSRNOT00000086650
ENSRNOT00000083645 |
Runx3
|
runt-related transcription factor 3 |
| chr9_-_81566642 | 0.02 |
ENSRNOT00000080345
|
Aamp
|
angio-associated, migratory cell protein |
| chr5_-_151895016 | 0.02 |
ENSRNOT00000081841
|
Pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr5_+_106952082 | 0.02 |
ENSRNOT00000046931
|
RGD1561246
|
similar to put. precursor MuIFN-alpha 5 |
| chr2_-_130658202 | 0.02 |
ENSRNOT00000017012
|
Pabpc4l
|
poly(A) binding protein, cytoplasmic 4-like |
| chr10_-_14937336 | 0.02 |
ENSRNOT00000025494
|
Sox8
|
SRY box 8 |
| chr5_+_133620706 | 0.02 |
ENSRNOT00000053202
|
Gm12830
|
predicted gene 12830 |
| chr17_+_82066152 | 0.02 |
ENSRNOT00000083034
|
Arl5b
|
ADP-ribosylation factor like GTPase 5B |
| chr11_+_45751812 | 0.02 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr13_-_70625842 | 0.02 |
ENSRNOT00000092499
|
Lamc2
|
laminin subunit gamma 2 |
| chr2_-_24923128 | 0.02 |
ENSRNOT00000044087
|
Pde8b
|
phosphodiesterase 8B |
| chrX_-_112328642 | 0.02 |
ENSRNOT00000083150
|
Psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr10_+_82937971 | 0.02 |
ENSRNOT00000005797
|
Dlx3
|
distal-less homeobox 3 |
| chr3_+_110367939 | 0.02 |
ENSRNOT00000010406
|
Bub1b
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr2_-_188553289 | 0.02 |
ENSRNOT00000088822
|
Trim46
|
tripartite motif-containing 46 |
| chr5_-_58113553 | 0.02 |
ENSRNOT00000046589
|
Arid3c
|
AT-rich interaction domain 3C |
| chr1_-_89045586 | 0.02 |
ENSRNOT00000063808
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr4_+_157513414 | 0.02 |
ENSRNOT00000078769
|
Pianp
|
PILR alpha associated neural protein |
| chr1_+_84044551 | 0.02 |
ENSRNOT00000028290
|
Coq8b
|
coenzyme Q8B |
| chr7_-_2961873 | 0.02 |
ENSRNOT00000067441
|
Rps15-ps2
|
ribosomal protein S15, pseudogene 2 |
| chr20_+_5125679 | 0.02 |
ENSRNOT00000060832
|
Bag6
|
BCL2-associated athanogene 6 |
| chr10_+_105787935 | 0.02 |
ENSRNOT00000000266
|
Mettl23
|
methyltransferase like 23 |
| chr4_+_180887182 | 0.02 |
ENSRNOT00000042270
ENSRNOT00000002476 |
Fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr2_-_28003186 | 0.02 |
ENSRNOT00000048554
|
Hexb
|
hexosaminidase subunit beta |
| chr1_-_220746224 | 0.02 |
ENSRNOT00000027734
|
Banf1
|
barrier to autointegration factor 1 |
| chr10_-_56412544 | 0.02 |
ENSRNOT00000020578
|
Tmem102
|
transmembrane protein 102 |
| chr1_+_219144205 | 0.02 |
ENSRNOT00000083942
|
Unc93b1
|
unc-93 homolog B1 (C. elegans) |
| chr3_-_146470293 | 0.02 |
ENSRNOT00000009627
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chrX_+_28435507 | 0.02 |
ENSRNOT00000005615
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chrX_-_18096938 | 0.02 |
ENSRNOT00000042670
|
LOC100912524
|
spindlin-2B-like |
| chr10_+_65448950 | 0.02 |
ENSRNOT00000082348
ENSRNOT00000037016 |
Rab34
|
RAB34, member RAS oncogene family |
| chr20_+_14577166 | 0.02 |
ENSRNOT00000085583
|
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr1_+_123015746 | 0.02 |
ENSRNOT00000013483
|
Magel2
|
MAGE family member L2 |
| chr5_+_140979435 | 0.02 |
ENSRNOT00000056592
|
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr10_+_65507364 | 0.01 |
ENSRNOT00000016297
|
Sdf2
|
stromal cell derived factor 2 |
| chr1_-_72727112 | 0.01 |
ENSRNOT00000031172
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr20_-_11737050 | 0.01 |
ENSRNOT00000001637
|
Sumo3
|
small ubiquitin-like modifier 3 |
| chr19_+_61677542 | 0.01 |
ENSRNOT00000014785
|
Itgb1
|
integrin subunit beta 1 |
| chr1_+_261291870 | 0.01 |
ENSRNOT00000049914
|
Hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr4_-_180887050 | 0.01 |
ENSRNOT00000065730
|
Ints13
|
integrator complex subunit 13 |
| chr10_+_20320878 | 0.01 |
ENSRNOT00000009714
|
Slit3
|
slit guidance ligand 3 |
| chr6_+_128750795 | 0.01 |
ENSRNOT00000005781
|
Glrx5
|
glutaredoxin 5 |
| chr15_-_45524582 | 0.01 |
ENSRNOT00000081912
|
Gucy1b2
|
guanylate cyclase 1 soluble subunit beta 2 |
| chr1_+_151918571 | 0.01 |
ENSRNOT00000022342
|
Ctsc
|
cathepsin C |
| chr3_+_150055749 | 0.01 |
ENSRNOT00000055335
|
Actl10
|
actin-like 10 |
| chr10_-_58784374 | 0.01 |
ENSRNOT00000019919
|
Med31
|
mediator complex subunit 31 |
| chr3_-_165741967 | 0.01 |
ENSRNOT00000017063
|
Zfp64
|
zinc finger protein 64 |
| chr6_-_24985716 | 0.01 |
ENSRNOT00000010613
|
Galnt14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr7_-_136853957 | 0.01 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr10_+_104019888 | 0.01 |
ENSRNOT00000032016
|
Slc16a5
|
solute carrier family 16 member 5 |
| chr10_-_56506446 | 0.01 |
ENSRNOT00000021357
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr7_-_120026755 | 0.01 |
ENSRNOT00000077677
|
Card10
|
caspase recruitment domain family, member 10 |
| chr8_-_111965889 | 0.01 |
ENSRNOT00000032376
|
Bfsp2
|
beaded filament structural protein 2 |
| chrX_+_54734385 | 0.01 |
ENSRNOT00000005023
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr5_-_101166651 | 0.01 |
ENSRNOT00000078862
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr8_-_97115027 | 0.01 |
ENSRNOT00000018685
|
Ankrd34c
|
ankyrin repeat domain 34C |
| chr1_+_282265370 | 0.01 |
ENSRNOT00000015687
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr5_-_107388182 | 0.01 |
ENSRNOT00000058358
|
LOC100912356
|
interferon alpha-1-like |
| chr1_+_79899155 | 0.01 |
ENSRNOT00000078503
|
Mypop
|
Myb-related transcription factor, partner of profilin |
| chr12_+_22835019 | 0.01 |
ENSRNOT00000059530
|
Col26a1
|
collagen type XXVI alpha 1 chain |
| chr9_-_114619711 | 0.01 |
ENSRNOT00000050012
|
Mtcl1
|
microtubule crosslinking factor 1 |
| chr10_+_46840113 | 0.01 |
ENSRNOT00000086083
ENSRNOT00000079133 |
Myo15a
|
myosin XVA |
| chr7_-_120027026 | 0.01 |
ENSRNOT00000011215
|
Card10
|
caspase recruitment domain family, member 10 |
| chr1_+_101178104 | 0.01 |
ENSRNOT00000028072
|
Pth2
|
parathyroid hormone 2 |
| chr8_-_59239954 | 0.01 |
ENSRNOT00000016104
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr1_-_219144610 | 0.01 |
ENSRNOT00000023526
|
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr8_+_116708027 | 0.01 |
ENSRNOT00000047309
|
Actl11
|
actin-like 11 |
| chr4_+_160168297 | 0.01 |
ENSRNOT00000083226
|
Cracr2a
|
calcium release activated channel regulator 2A |
| chr6_-_24563246 | 0.01 |
ENSRNOT00000074294
|
LOC685881
|
hypothetical protein LOC685881 |
| chr7_+_28066635 | 0.01 |
ENSRNOT00000005844
|
Pah
|
phenylalanine hydroxylase |
| chr3_-_151363115 | 0.01 |
ENSRNOT00000073492
|
Eif6
|
eukaryotic translation initiation factor 6 |
| chr4_-_67520356 | 0.01 |
ENSRNOT00000014604
|
Braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr1_+_222198534 | 0.01 |
ENSRNOT00000028712
|
Bad
|
BCL2-associated agonist of cell death |
| chr5_+_68717519 | 0.01 |
ENSRNOT00000066226
|
Smc2
|
structural maintenance of chromosomes 2 |
| chr17_+_43460782 | 0.01 |
ENSRNOT00000059494
|
Slc17a4
|
solute carrier family 17, member 4 |
| chr10_-_85289777 | 0.01 |
ENSRNOT00000055427
ENSRNOT00000014863 |
Gpr179
|
G protein-coupled receptor 179 |
| chr7_-_143473697 | 0.01 |
ENSRNOT00000055336
ENSRNOT00000083027 |
Krt77
|
keratin 77 |
| chr18_+_16650806 | 0.01 |
ENSRNOT00000093679
ENSRNOT00000041961 ENSRNOT00000093140 |
Fhod3
|
formin homology 2 domain containing 3 |
| chr8_+_71719563 | 0.01 |
ENSRNOT00000022828
|
Ppib
|
peptidylprolyl isomerase B |
| chr8_+_85489553 | 0.01 |
ENSRNOT00000082158
|
Rpl12-ps1
|
ribosomal protein L12, pseudogene 1 |
| chr17_+_86199623 | 0.01 |
ENSRNOT00000022727
|
Ptf1a
|
pancreas specific transcription factor, 1a |
| chr9_+_99998275 | 0.01 |
ENSRNOT00000074395
|
Gpc1
|
glypican 1 |
| chr6_+_136380990 | 0.01 |
ENSRNOT00000063865
ENSRNOT00000082914 |
Zfyve21
|
zinc finger FYVE-type containing 21 |
| chr10_-_34439470 | 0.01 |
ENSRNOT00000072081
|
Btnl9
|
butyrophilin-like 9 |
| chr3_-_94767239 | 0.01 |
ENSRNOT00000056808
|
Qser1
|
glutamine and serine rich 1 |
| chr17_+_8135251 | 0.01 |
ENSRNOT00000016982
|
Trpc7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr2_+_243728434 | 0.01 |
ENSRNOT00000017252
|
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr2_+_240021152 | 0.01 |
ENSRNOT00000012473
|
Tacr3
|
tachykinin receptor 3 |
| chr12_+_39913628 | 0.01 |
ENSRNOT00000046604
|
Ccdc63
|
coiled-coil domain containing 63 |
| chr14_-_17436897 | 0.01 |
ENSRNOT00000003277
|
Uso1
|
USO1 vesicle transport factor |
| chr1_+_84304228 | 0.01 |
ENSRNOT00000024771
|
Prx
|
periaxin |
| chr2_-_21698937 | 0.01 |
ENSRNOT00000080165
|
AABR07007642.1
|
|
| chr4_+_99823252 | 0.01 |
ENSRNOT00000013587
|
Polr1a
|
RNA polymerase I subunit A |
| chr10_+_15620871 | 0.01 |
ENSRNOT00000089435
ENSRNOT00000066430 |
Nprl3
|
NPR3-like, GATOR1 complex subunit |
| chr12_+_31536951 | 0.01 |
ENSRNOT00000086078
|
Rimbp2
|
RIMS binding protein 2 |
| chr1_+_280423079 | 0.01 |
ENSRNOT00000011983
|
Slc18a2
|
solute carrier family 18 member A2 |
| chr3_-_176548208 | 0.01 |
ENSRNOT00000084339
|
Chrna4
|
cholinergic receptor nicotinic alpha 4 subunit |
| chr10_+_12980674 | 0.01 |
ENSRNOT00000004638
|
Bicdl2
|
BICD family like cargo adaptor 2 |
| chr3_+_35014538 | 0.01 |
ENSRNOT00000006341
|
Kif5c
|
kinesin family member 5C |
| chr2_-_218951030 | 0.01 |
ENSRNOT00000019172
|
Slc30a7
|
solute carrier family 30 member 7 |
| chr2_-_56679955 | 0.01 |
ENSRNOT00000016722
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr13_-_72890841 | 0.01 |
ENSRNOT00000093726
|
RGD1304622
|
similar to 6820428L09 protein |
| chr1_+_199248470 | 0.00 |
ENSRNOT00000025933
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr20_+_5442390 | 0.00 |
ENSRNOT00000037499
|
Rps18
|
ribosomal protein S18 |
| chr1_+_282568287 | 0.00 |
ENSRNOT00000015997
|
Ces2i
|
carboxylesterase 2I |
| chr15_+_344360 | 0.00 |
ENSRNOT00000077671
|
Kcnma1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr1_-_187779675 | 0.00 |
ENSRNOT00000024648
|
Arl6ip1
|
ADP-ribosylation factor like GTPase 6 interacting protein 1 |
| chr10_+_56237755 | 0.00 |
ENSRNOT00000016163
|
Fxr2
|
FMR1 autosomal homolog 2 |
| chr2_-_218658761 | 0.00 |
ENSRNOT00000018318
|
S1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr1_-_216971183 | 0.00 |
ENSRNOT00000077911
|
Mrgpre
|
MAS related GPR family member E |
| chr1_-_65681440 | 0.00 |
ENSRNOT00000026305
|
Zfp128
|
zinc finger protein 128 |
| chr2_+_27365148 | 0.00 |
ENSRNOT00000021549
|
Col4a3bp
|
collagen type IV alpha 3 binding protein |
| chr3_+_8534440 | 0.00 |
ENSRNOT00000045827
ENSRNOT00000082672 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr1_+_260093641 | 0.00 |
ENSRNOT00000019521
|
Ccnj
|
cyclin J |
| chr3_-_11317049 | 0.00 |
ENSRNOT00000030558
|
Swi5
|
SWI5 homologous recombination repair protein |
| chr3_-_14019204 | 0.00 |
ENSRNOT00000072400
ENSRNOT00000092918 |
Traf1
|
TNF receptor-associated factor 1 |
| chr6_-_43448280 | 0.00 |
ENSRNOT00000081110
|
Adam17
|
ADAM metallopeptidase domain 17 |
| chr18_+_30398113 | 0.00 |
ENSRNOT00000027206
|
Pcdhb5
|
protocadherin beta 5 |
| chr18_-_81807627 | 0.00 |
ENSRNOT00000020376
|
Timm21
|
translocase of inner mitochondrial membrane 21 |
| chr14_+_13192347 | 0.00 |
ENSRNOT00000000092
|
Antxr2
|
anthrax toxin receptor 2 |
| chr3_-_2431148 | 0.00 |
ENSRNOT00000012978
|
Nelfb
|
negative elongation factor complex member B |
| chr10_-_35833861 | 0.00 |
ENSRNOT00000049942
|
Canx
|
calnexin |
| chr7_-_70476340 | 0.00 |
ENSRNOT00000006800
|
Arhgef25
|
Rho guanine nucleotide exchange factor 25 |
| chr8_-_55681930 | 0.00 |
ENSRNOT00000065744
|
Colca2
|
colorectal cancer associated 2 |
| chr1_+_72635267 | 0.00 |
ENSRNOT00000090636
|
Il11
|
interleukin 11 |
| chr4_-_132111079 | 0.00 |
ENSRNOT00000013719
|
Eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr3_+_72447969 | 0.00 |
ENSRNOT00000012022
|
Ssrp1
|
structure specific recognition protein 1 |
| chr1_+_245396989 | 0.00 |
ENSRNOT00000016757
|
Kcnv2
|
potassium voltage-gated channel modifier subfamily V member 2 |
| chr1_+_192169841 | 0.00 |
ENSRNOT00000025832
|
Chp2
|
calcineurin-like EF hand protein 2 |
| chr3_+_79860179 | 0.00 |
ENSRNOT00000081160
ENSRNOT00000068444 |
Rapsn
|
receptor-associated protein of the synapse |
| chr3_-_161212188 | 0.00 |
ENSRNOT00000065751
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr3_-_93734282 | 0.00 |
ENSRNOT00000012428
|
Caprin1
|
cell cycle associated protein 1 |
| chr5_-_74190991 | 0.00 |
ENSRNOT00000090366
|
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr20_+_5125349 | 0.00 |
ENSRNOT00000085598
ENSRNOT00000001129 |
Bag6
|
BCL2-associated athanogene 6 |
| chr16_-_74864816 | 0.00 |
ENSRNOT00000017164
|
Alg11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr4_-_6062641 | 0.00 |
ENSRNOT00000074846
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chrX_+_65226748 | 0.00 |
ENSRNOT00000076181
|
Msn
|
moesin |
| chr16_-_5795825 | 0.00 |
ENSRNOT00000048043
|
Cacna2d3
|
calcium voltage-gated channel auxiliary subunit alpha2delta 3 |
| chr6_+_105364668 | 0.00 |
ENSRNOT00000009513
ENSRNOT00000087090 |
Ttc9
|
tetratricopeptide repeat domain 9 |
| chr2_+_104416972 | 0.00 |
ENSRNOT00000017125
|
Trim55
|
tripartite motif-containing 55 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zic3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:2000546 | positive regulation of fat cell proliferation(GO:0070346) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |