Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
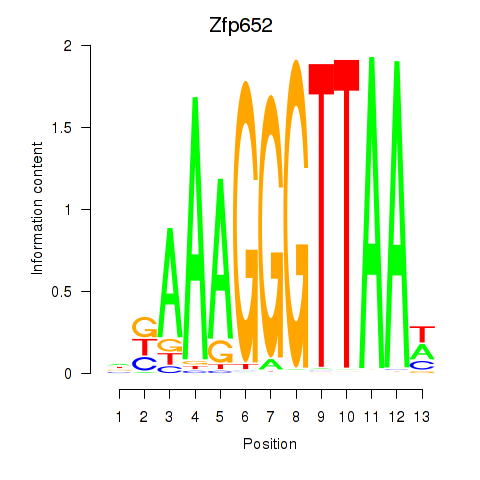
Results for Zfp652
Z-value: 0.16
Transcription factors associated with Zfp652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp652
|
ENSRNOG00000017001 | zinc finger protein 652 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp652 | rn6_v1_chr10_+_83556809_83556809 | 0.87 | 5.6e-02 | Click! |
Activity profile of Zfp652 motif
Sorted Z-values of Zfp652 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_66417741 | 0.13 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr10_-_82852660 | 0.09 |
ENSRNOT00000005641
|
Pdk2
|
pyruvate dehydrogenase kinase 2 |
| chr1_-_43638161 | 0.05 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr7_-_123088819 | 0.05 |
ENSRNOT00000056077
|
AC096601.1
|
|
| chr6_-_42473738 | 0.04 |
ENSRNOT00000033327
|
Kcnf1
|
potassium voltage-gated channel modifier subfamily F member 1 |
| chr6_+_119519714 | 0.04 |
ENSRNOT00000004955
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr5_-_59638385 | 0.04 |
ENSRNOT00000060203
ENSRNOT00000077367 |
Rnf38
|
ring finger protein 38 |
| chr5_+_79055521 | 0.04 |
ENSRNOT00000010333
|
Col27a1
|
collagen type XXVII alpha 1 chain |
| chr11_-_71695000 | 0.04 |
ENSRNOT00000073550
|
Ubxn7
|
UBX domain protein 7 |
| chr16_-_79973735 | 0.03 |
ENSRNOT00000057845
ENSRNOT00000086896 |
Dlgap2
|
DLG associated protein 2 |
| chr6_+_99625306 | 0.03 |
ENSRNOT00000008573
|
Plekhg3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr13_+_71107465 | 0.03 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr2_+_50099576 | 0.03 |
ENSRNOT00000089218
|
Hcn1
|
hyperpolarization-activated cyclic nucleotide-gated potassium channel 1 |
| chr1_-_239057732 | 0.03 |
ENSRNOT00000024775
|
Gda
|
guanine deaminase |
| chr11_+_57404196 | 0.03 |
ENSRNOT00000042754
ENSRNOT00000034097 |
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr15_-_28746042 | 0.03 |
ENSRNOT00000017730
|
Sall2
|
spalt-like transcription factor 2 |
| chr7_-_14402010 | 0.03 |
ENSRNOT00000008303
|
Wiz
|
widely-interspaced zinc finger motifs |
| chr4_+_7355574 | 0.03 |
ENSRNOT00000013800
|
Kcnh2
|
potassium voltage-gated channel subfamily H member 2 |
| chr6_-_102196138 | 0.03 |
ENSRNOT00000014132
|
Tmem229b
|
transmembrane protein 229B |
| chr3_-_117990289 | 0.03 |
ENSRNOT00000011084
|
Shc4
|
SHC adaptor protein 4 |
| chr8_-_78655856 | 0.02 |
ENSRNOT00000081185
|
Tcf12
|
transcription factor 12 |
| chr3_-_11452529 | 0.02 |
ENSRNOT00000020206
|
Slc25a25
|
solute carrier family 25 member 25 |
| chr5_-_79222687 | 0.02 |
ENSRNOT00000010516
|
Akna
|
AT-hook transcription factor |
| chr1_-_199064150 | 0.02 |
ENSRNOT00000044511
|
Zfp689
|
zinc finger protein 689 |
| chr10_+_84955864 | 0.02 |
ENSRNOT00000038572
|
Sp6
|
Sp6 transcription factor |
| chr10_-_109224370 | 0.02 |
ENSRNOT00000005839
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr5_+_58144705 | 0.02 |
ENSRNOT00000019886
|
Galt
|
galactose-1-phosphate uridylyltransferase |
| chr20_-_7307280 | 0.02 |
ENSRNOT00000000583
ENSRNOT00000079933 |
Spdef
|
SAM pointed domain containing ets transcription factor |
| chr7_+_78558701 | 0.02 |
ENSRNOT00000006393
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr8_+_22189600 | 0.02 |
ENSRNOT00000061100
|
Pde4a
|
phosphodiesterase 4A |
| chr3_+_45277348 | 0.02 |
ENSRNOT00000059246
|
Pkp4
|
plakophilin 4 |
| chr10_-_86645529 | 0.01 |
ENSRNOT00000011947
|
Med24
|
mediator complex subunit 24 |
| chr1_-_84254645 | 0.01 |
ENSRNOT00000077651
|
Sptbn4
|
spectrin, beta, non-erythrocytic 4 |
| chr6_-_51018050 | 0.01 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chr3_-_72289310 | 0.01 |
ENSRNOT00000038250
|
Rtn4rl2
|
reticulon 4 receptor-like 2 |
| chr17_-_15467320 | 0.01 |
ENSRNOT00000072490
ENSRNOT00000093561 |
Ogn
|
osteoglycin |
| chr11_-_14741563 | 0.01 |
ENSRNOT00000002152
|
Nrip1
|
nuclear receptor interacting protein 1 |
| chr3_-_162581610 | 0.01 |
ENSRNOT00000079324
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr5_-_152473868 | 0.01 |
ENSRNOT00000022130
|
Fam110d
|
family with sequence similarity 110, member D |
| chr5_+_60250546 | 0.01 |
ENSRNOT00000017707
|
Zcchc7
|
zinc finger CCHC-type containing 7 |
| chr10_+_53466870 | 0.01 |
ENSRNOT00000057503
|
Pirt
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr8_+_117648745 | 0.01 |
ENSRNOT00000027762
|
Slc26a6
|
solute carrier family 26 member 6 |
| chr15_-_58171049 | 0.01 |
ENSRNOT00000001371
|
Gpalpp1
|
GPALPP motifs containing 1 |
| chr12_-_31249118 | 0.01 |
ENSRNOT00000058586
ENSRNOT00000073105 |
Adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr2_-_219090874 | 0.01 |
ENSRNOT00000019377
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chr3_-_138116664 | 0.01 |
ENSRNOT00000055602
|
Rrbp1
|
ribosome binding protein 1 |
| chr5_-_152589719 | 0.01 |
ENSRNOT00000022522
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr7_-_140291620 | 0.01 |
ENSRNOT00000088323
|
Adcy6
|
adenylate cyclase 6 |
| chr7_+_2635743 | 0.01 |
ENSRNOT00000004223
|
Mip
|
major intrinsic protein of lens fiber |
| chr10_+_56627411 | 0.01 |
ENSRNOT00000089108
ENSRNOT00000068493 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr9_+_93326283 | 0.01 |
ENSRNOT00000024578
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr1_+_157920786 | 0.01 |
ENSRNOT00000014284
|
Fam181b
|
family with sequence similarity 181, member B |
| chr9_-_69878706 | 0.01 |
ENSRNOT00000035879
|
Ino80d
|
INO80 complex subunit D |
| chrX_-_62690806 | 0.01 |
ENSRNOT00000018147
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr13_-_88436789 | 0.01 |
ENSRNOT00000003901
|
Ddr2
|
discoidin domain receptor tyrosine kinase 2 |
| chr8_-_61823102 | 0.01 |
ENSRNOT00000058645
|
Neil1
|
nei-like DNA glycosylase 1 |
| chr6_-_108596446 | 0.01 |
ENSRNOT00000031331
ENSRNOT00000077458 ENSRNOT00000082792 |
Ltbp2
|
latent transforming growth factor beta binding protein 2 |
| chr13_+_60435946 | 0.00 |
ENSRNOT00000004342
|
B3galt2
|
Beta-1,3-galactosyltransferase 2 |
| chr8_-_63350269 | 0.00 |
ENSRNOT00000042681
|
Cd276
|
Cd276 molecule |
| chr18_-_61057365 | 0.00 |
ENSRNOT00000042352
|
Alpk2
|
alpha-kinase 2 |
| chr1_-_84040433 | 0.00 |
ENSRNOT00000018696
|
Itpkc
|
inositol-trisphosphate 3-kinase C |
| chr7_-_117259141 | 0.00 |
ENSRNOT00000040762
|
Plec
|
plectin |
| chr10_+_31146107 | 0.00 |
ENSRNOT00000008184
|
Adam19
|
ADAM metallopeptidase domain 19 |
| chr11_+_74834050 | 0.00 |
ENSRNOT00000002333
|
Atp13a4
|
ATPase 13A4 |
| chr8_+_63600663 | 0.00 |
ENSRNOT00000012644
|
Hcn4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr1_+_79989019 | 0.00 |
ENSRNOT00000020428
|
Dmpk
|
dystrophia myotonica-protein kinase |
| chr10_+_91200874 | 0.00 |
ENSRNOT00000004272
|
Hexim1
|
hexamethylene bis-acetamide inducible 1 |
| chr15_-_60766579 | 0.00 |
ENSRNOT00000079978
|
Akap11
|
A-kinase anchoring protein 11 |
| chr1_-_271275989 | 0.00 |
ENSRNOT00000075570
|
LOC100912218
|
inositol 1,4,5-trisphosphate receptor-interacting protein-like |
| chr11_-_87017115 | 0.00 |
ENSRNOT00000051037
|
Rtn4r
|
reticulon 4 receptor |
| chr2_-_207300854 | 0.00 |
ENSRNOT00000018061
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr17_-_14717420 | 0.00 |
ENSRNOT00000071131
|
LOC100910978
|
extracellular matrix protein 2-like |
| chr17_-_15555919 | 0.00 |
ENSRNOT00000043852
|
Ecm2
|
extracellular matrix protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp652
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |