Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
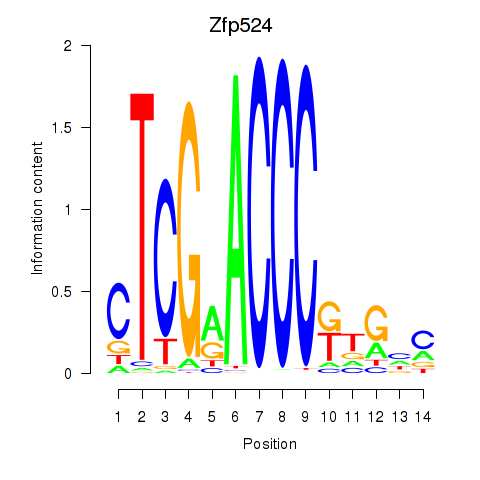
Results for Zfp524
Z-value: 0.70
Transcription factors associated with Zfp524
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp524
|
ENSRNOG00000016565 | zinc finger protein 524 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp524 | rn6_v1_chr1_-_72377434_72377434 | -0.58 | 3.0e-01 | Click! |
Activity profile of Zfp524 motif
Sorted Z-values of Zfp524 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_89008008 | 0.50 |
ENSRNOT00000074586
|
Cebpd
|
CCAAT/enhancer binding protein delta |
| chr10_-_97582188 | 0.46 |
ENSRNOT00000005076
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr16_+_20352480 | 0.38 |
ENSRNOT00000025956
|
Arrdc2
|
arrestin domain containing 2 |
| chr16_-_8685529 | 0.34 |
ENSRNOT00000092751
|
Slc18a3
|
solute carrier family 18 member A3 |
| chr18_+_63130542 | 0.28 |
ENSRNOT00000024947
|
Tubb6
|
tubulin, beta 6 class V |
| chr13_-_50499060 | 0.27 |
ENSRNOT00000065347
ENSRNOT00000076924 |
Etnk2
|
ethanolamine kinase 2 |
| chr4_-_28702559 | 0.26 |
ENSRNOT00000013910
ENSRNOT00000013937 |
Calcr
|
calcitonin receptor |
| chr2_-_257484587 | 0.25 |
ENSRNOT00000083344
|
Nexn
|
nexilin (F actin binding protein) |
| chr9_-_5330815 | 0.25 |
ENSRNOT00000014548
|
Slc5a7
|
solute carrier family 5 member 7 |
| chr1_+_205842489 | 0.24 |
ENSRNOT00000081610
|
Fank1
|
fibronectin type 3 and ankyrin repeat domains 1 |
| chr11_+_67465236 | 0.24 |
ENSRNOT00000042374
|
Stfa2
|
stefin A2 |
| chr7_+_141370491 | 0.24 |
ENSRNOT00000087662
|
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 |
| chr19_-_37427989 | 0.24 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr19_-_11308740 | 0.23 |
ENSRNOT00000067391
|
Mt2A
|
metallothionein 2A |
| chr5_-_127983515 | 0.22 |
ENSRNOT00000043054
|
Fam159a
|
family with sequence similarity 159, member A |
| chr12_+_47698947 | 0.22 |
ENSRNOT00000001586
|
Trpv4
|
transient receptor potential cation channel, subfamily V, member 4 |
| chr13_+_52588917 | 0.22 |
ENSRNOT00000011999
|
Phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr1_+_279798187 | 0.22 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr12_-_22477052 | 0.21 |
ENSRNOT00000075504
|
Ache
|
acetylcholinesterase |
| chr9_-_82146874 | 0.20 |
ENSRNOT00000024190
|
Fev
|
FEV, ETS transcription factor |
| chr17_+_24416651 | 0.19 |
ENSRNOT00000024458
|
Cd83
|
CD83 molecule |
| chr1_-_88881460 | 0.19 |
ENSRNOT00000028287
|
Hcst
|
hematopoietic cell signal transducer |
| chr13_-_44345735 | 0.19 |
ENSRNOT00000005006
|
Tmem163
|
transmembrane protein 163 |
| chr3_-_111422203 | 0.18 |
ENSRNOT00000084290
|
Oip5
|
Opa interacting protein 5 |
| chr1_+_163445527 | 0.18 |
ENSRNOT00000020520
|
Lrrc32
|
leucine rich repeat containing 32 |
| chr13_+_89386023 | 0.18 |
ENSRNOT00000086223
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr1_+_198690794 | 0.18 |
ENSRNOT00000023999
|
Zfp771
|
zinc finger protein 771 |
| chr11_+_64882288 | 0.18 |
ENSRNOT00000077727
|
Pla1a
|
phospholipase A1 member A |
| chr13_+_89385859 | 0.18 |
ENSRNOT00000047434
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr19_+_44164935 | 0.18 |
ENSRNOT00000048998
|
Gabarapl2
|
GABA type A receptor associated protein like 2 |
| chr4_-_10269748 | 0.17 |
ENSRNOT00000074662
|
Fam185a
|
family with sequence similarity 185, member A |
| chr20_+_32450733 | 0.17 |
ENSRNOT00000036449
|
Rsph4a
|
radial spoke head 4 homolog A |
| chr11_+_47243342 | 0.16 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr10_+_86303727 | 0.16 |
ENSRNOT00000037752
|
Ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr1_+_238222521 | 0.16 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr11_+_32211115 | 0.16 |
ENSRNOT00000087452
|
Mrps6
|
mitochondrial ribosomal protein S6 |
| chr11_+_87204175 | 0.16 |
ENSRNOT00000000306
|
Slc25a1
|
solute carrier family 25 member 1 |
| chr2_-_200513564 | 0.16 |
ENSRNOT00000056173
|
Phgdh
|
phosphoglycerate dehydrogenase |
| chr5_-_143062226 | 0.15 |
ENSRNOT00000029388
|
Dnali1
|
dynein, axonemal, light intermediate chain 1 |
| chr10_-_54512169 | 0.15 |
ENSRNOT00000005066
|
Cfap52
|
cilia and flagella associated protein 52 |
| chr19_+_25123724 | 0.15 |
ENSRNOT00000007407
|
LOC686013
|
hypothetical protein LOC686013 |
| chr7_+_123573930 | 0.14 |
ENSRNOT00000066149
|
Fam109b
|
family with sequence similarity 109, member B |
| chr20_+_33945829 | 0.14 |
ENSRNOT00000064063
|
LOC103689994
|
radial spoke head protein 4 homolog A |
| chr10_+_3218466 | 0.14 |
ENSRNOT00000093629
ENSRNOT00000093338 ENSRNOT00000077695 |
Ntan1
|
N-terminal asparagine amidase |
| chr1_-_212548730 | 0.14 |
ENSRNOT00000089729
|
Caly
|
calcyon neuron-specific vesicular protein |
| chr17_+_9736577 | 0.14 |
ENSRNOT00000066586
|
F12
|
coagulation factor XII |
| chr15_+_24141651 | 0.14 |
ENSRNOT00000082304
|
Lgals3
|
galectin 3 |
| chr2_+_127538659 | 0.14 |
ENSRNOT00000093483
ENSRNOT00000058476 |
Slc25a31
|
solute carrier family 25 member 31 |
| chr2_-_188413219 | 0.14 |
ENSRNOT00000065065
|
Fdps
|
farnesyl diphosphate synthase |
| chr4_+_145580799 | 0.13 |
ENSRNOT00000013727
|
Vhl
|
von Hippel-Lindau tumor suppressor |
| chr8_+_118378059 | 0.13 |
ENSRNOT00000043247
|
AABR07071482.1
|
|
| chr8_-_82351108 | 0.13 |
ENSRNOT00000013053
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr11_-_36479868 | 0.13 |
ENSRNOT00000075762
|
LOC100911295
|
non-histone chromosomal protein HMG-14-like |
| chr6_-_45669148 | 0.13 |
ENSRNOT00000010092
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr6_+_2969333 | 0.13 |
ENSRNOT00000047356
|
Morn2
|
MORN repeat containing 2 |
| chr6_-_91518996 | 0.13 |
ENSRNOT00000005835
|
Pole2
|
DNA polymerase epsilon 2, accessory subunit |
| chr7_+_117394372 | 0.13 |
ENSRNOT00000048706
|
Gpaa1
|
glycosylphosphatidylinositol anchor attachment 1 |
| chr3_+_72385666 | 0.13 |
ENSRNOT00000011168
|
Prg2
|
proteoglycan 2 |
| chr16_-_61791091 | 0.13 |
ENSRNOT00000042003
|
Mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr5_-_58106706 | 0.13 |
ENSRNOT00000019561
|
Dctn3
|
dynactin subunit 3 |
| chr4_+_62780596 | 0.13 |
ENSRNOT00000073846
|
LOC689574
|
hypothetical protein LOC689574 |
| chr5_-_75676584 | 0.12 |
ENSRNOT00000044348
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr17_+_15845931 | 0.12 |
ENSRNOT00000092083
|
Card19
|
caspase recruitment domain family, member 19 |
| chr1_-_43884267 | 0.12 |
ENSRNOT00000024418
|
Cnksr3
|
Cnksr family member 3 |
| chr3_+_72226613 | 0.12 |
ENSRNOT00000010619
|
Timm10
|
translocase of inner mitochondrial membrane 10 |
| chr10_+_85646633 | 0.12 |
ENSRNOT00000086608
|
Psmb3
|
proteasome subunit beta 3 |
| chr11_-_32088002 | 0.12 |
ENSRNOT00000002732
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr10_+_15241590 | 0.12 |
ENSRNOT00000037372
ENSRNOT00000037381 |
Mettl26
|
methyltransferase like 26 |
| chr13_+_83681322 | 0.12 |
ENSRNOT00000004206
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr9_+_100076958 | 0.12 |
ENSRNOT00000084297
|
Dusp28
|
dual specificity phosphatase 28 |
| chr20_-_3819200 | 0.12 |
ENSRNOT00000000542
ENSRNOT00000081486 |
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr15_+_33121273 | 0.12 |
ENSRNOT00000016020
|
Rem2
|
RRAD and GEM like GTPase 2 |
| chr10_-_58924137 | 0.11 |
ENSRNOT00000066740
|
Tekt1
|
tektin 1 |
| chr1_+_31967978 | 0.11 |
ENSRNOT00000081471
ENSRNOT00000021532 |
Trip13
|
thyroid hormone receptor interactor 13 |
| chr1_-_190914610 | 0.11 |
ENSRNOT00000023189
|
Cdr2
|
cerebellar degeneration-related protein 2 |
| chr3_-_171342646 | 0.11 |
ENSRNOT00000071853
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr17_+_70684340 | 0.11 |
ENSRNOT00000051067
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr16_+_80826681 | 0.11 |
ENSRNOT00000000123
|
Coprs
|
coordinator of PRMT5 and differentiation stimulator |
| chr1_-_89474252 | 0.11 |
ENSRNOT00000028597
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr3_-_7632345 | 0.11 |
ENSRNOT00000049889
|
Cfap77
|
cilia and flagella associated protein 77 |
| chr7_-_143353925 | 0.11 |
ENSRNOT00000068533
|
Krt71
|
keratin 71 |
| chr2_-_189573280 | 0.11 |
ENSRNOT00000022897
|
Rps27
|
ribosomal protein S27 |
| chr19_+_37226186 | 0.11 |
ENSRNOT00000075933
ENSRNOT00000065013 |
Hsf4
|
heat shock transcription factor 4 |
| chr13_+_88557860 | 0.11 |
ENSRNOT00000058547
|
Sh2d1b2
|
SH2 domain containing 1B2 |
| chr5_-_167999853 | 0.11 |
ENSRNOT00000087402
|
Park7
|
Parkinsonism associated deglycase |
| chr1_+_202432366 | 0.11 |
ENSRNOT00000027681
|
Plpp4
|
phospholipid phosphatase 4 |
| chr16_+_10250404 | 0.11 |
ENSRNOT00000087037
ENSRNOT00000081183 |
Gdf10
|
growth differentiation factor 10 |
| chr3_+_23301455 | 0.11 |
ENSRNOT00000019405
|
Arpc5l
|
actin related protein 2/3 complex, subunit 5-like |
| chr19_+_24800072 | 0.11 |
ENSRNOT00000005470
|
Ptger1
|
prostaglandin E receptor 1 |
| chr4_+_112773938 | 0.10 |
ENSRNOT00000009321
|
Eva1a
|
eva-1 homolog A, regulator of programmed cell death |
| chr11_+_30363280 | 0.10 |
ENSRNOT00000002885
|
Sod1
|
superoxide dismutase 1, soluble |
| chr1_-_173764246 | 0.10 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chr17_+_18031228 | 0.10 |
ENSRNOT00000081708
|
Tpmt
|
thiopurine S-methyltransferase |
| chr6_-_27024129 | 0.10 |
ENSRNOT00000012273
|
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr1_+_87248489 | 0.10 |
ENSRNOT00000028091
|
Dpf1
|
double PHD fingers 1 |
| chr15_-_19575914 | 0.10 |
ENSRNOT00000043897
|
AABR07017253.1
|
|
| chr13_+_51972974 | 0.10 |
ENSRNOT00000008164
|
Arl8a
|
ADP-ribosylation factor like GTPase 8A |
| chr9_-_17698569 | 0.10 |
ENSRNOT00000087528
|
Mrpl14
|
mitochondrial ribosomal protein L14 |
| chr10_+_86340940 | 0.10 |
ENSRNOT00000073486
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr20_-_5192000 | 0.10 |
ENSRNOT00000079677
|
Tnf
|
tumor necrosis factor |
| chr16_-_74408030 | 0.10 |
ENSRNOT00000026418
|
Slc20a2
|
solute carrier family 20 member 2 |
| chr19_+_24456976 | 0.10 |
ENSRNOT00000004900
|
Ucp1
|
uncoupling protein 1 |
| chr17_-_2705123 | 0.09 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr2_-_211017778 | 0.09 |
ENSRNOT00000026883
|
Sypl2
|
synaptophysin-like 2 |
| chr9_+_17784468 | 0.09 |
ENSRNOT00000026831
|
Slc29a1
|
solute carrier family 29 member 1 |
| chr19_+_21372163 | 0.09 |
ENSRNOT00000020329
|
Siah1
|
siah E3 ubiquitin protein ligase 1 |
| chrX_-_20807216 | 0.09 |
ENSRNOT00000075757
|
Fam156b
|
family with sequence similarity 156, member B |
| chr20_-_1984737 | 0.09 |
ENSRNOT00000040232
ENSRNOT00000051634 ENSRNOT00000079445 |
Gabbr1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr17_+_16333415 | 0.09 |
ENSRNOT00000060550
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr11_-_61530567 | 0.09 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr5_-_145392123 | 0.09 |
ENSRNOT00000019266
|
Gjb3
|
gap junction protein, beta 3 |
| chr19_+_16417817 | 0.09 |
ENSRNOT00000015272
|
Irx5
|
iroquois homeobox 5 |
| chr6_+_111296417 | 0.09 |
ENSRNOT00000075425
|
Ahsa1
|
activator of Hsp90 ATPase activity 1 |
| chr2_+_204932159 | 0.09 |
ENSRNOT00000078376
|
Ngf
|
nerve growth factor |
| chr10_+_14598014 | 0.09 |
ENSRNOT00000068336
|
Tsr3
|
TSR3, 20S rRNA accumulation |
| chr19_-_19376789 | 0.09 |
ENSRNOT00000065103
|
Nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr20_+_4855829 | 0.09 |
ENSRNOT00000001110
|
LOC103694380
|
tumor necrosis factor-like |
| chr17_-_53102342 | 0.09 |
ENSRNOT00000073834
|
Psma2
|
proteasome subunit alpha 2 |
| chr4_+_78735279 | 0.09 |
ENSRNOT00000011970
|
Malsu1
|
mitochondrial assembly of ribosomal large subunit 1 |
| chr17_-_62299655 | 0.09 |
ENSRNOT00000024855
|
Gjd4
|
gap junction protein, delta 4 |
| chr5_+_14890408 | 0.09 |
ENSRNOT00000033116
|
Sox17
|
SRY box 17 |
| chr10_+_64556064 | 0.09 |
ENSRNOT00000010778
|
Timm22
|
translocase of inner mitochondrial membrane 22 |
| chr6_+_75429729 | 0.09 |
ENSRNOT00000043250
|
Rps10l1
|
ribosomal protein S10-like 1 |
| chr7_-_67345295 | 0.09 |
ENSRNOT00000087045
ENSRNOT00000005829 |
Avpr1a
|
arginine vasopressin receptor 1A |
| chr3_+_160908769 | 0.09 |
ENSRNOT00000030054
|
Sys1
|
Sys1 golgi trafficking protein |
| chr13_+_82298534 | 0.09 |
ENSRNOT00000003636
|
Mettl18
|
methyltransferase like 18 |
| chr4_+_171748273 | 0.09 |
ENSRNOT00000009998
|
Dera
|
deoxyribose-phosphate aldolase |
| chr13_+_49074644 | 0.09 |
ENSRNOT00000000041
|
Klhdc8a
|
kelch domain containing 8A |
| chr20_-_10842370 | 0.09 |
ENSRNOT00000001580
|
Hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr12_-_18540166 | 0.09 |
ENSRNOT00000001792
|
Il3ra
|
interleukin 3 receptor subunit alpha |
| chr1_-_101514547 | 0.09 |
ENSRNOT00000079633
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr2_-_39434560 | 0.09 |
ENSRNOT00000090193
|
Ndufaf2
|
NADH:ubiquinone oxidoreductase complex assembly factor 2 |
| chr3_+_8450612 | 0.09 |
ENSRNOT00000040457
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr9_-_99818262 | 0.09 |
ENSRNOT00000056600
|
Cops9
|
COP9 signalosome subunit 9 |
| chr8_+_21663325 | 0.09 |
ENSRNOT00000027749
|
Ubl5
|
ubiquitin-like 5 |
| chr1_+_16910069 | 0.08 |
ENSRNOT00000020015
|
Aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr3_-_81282157 | 0.08 |
ENSRNOT00000051258
|
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr2_-_143104412 | 0.08 |
ENSRNOT00000058116
|
Ufm1
|
ubiquitin-fold modifier 1 |
| chr1_-_52894832 | 0.08 |
ENSRNOT00000016471
|
T
|
T brachyury transcription factor |
| chr8_+_119135013 | 0.08 |
ENSRNOT00000056114
|
Prss50
|
protease, serine, 50 |
| chr9_+_43259709 | 0.08 |
ENSRNOT00000022487
|
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr12_+_37211316 | 0.08 |
ENSRNOT00000032739
|
Ccdc92
|
coiled-coil domain containing 92 |
| chr6_+_8346704 | 0.08 |
ENSRNOT00000092218
|
AC111831.2
|
|
| chr20_+_3744395 | 0.08 |
ENSRNOT00000086065
|
Tcf19
|
transcription factor 19 |
| chr11_+_64790801 | 0.08 |
ENSRNOT00000004023
|
Timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr16_-_8686131 | 0.08 |
ENSRNOT00000080202
|
Chat
|
choline O-acetyltransferase |
| chr5_+_172648950 | 0.08 |
ENSRNOT00000055361
|
Faap20
|
Fanconi anemia core complex associated protein 20 |
| chrX_+_26294066 | 0.08 |
ENSRNOT00000037862
|
Hccs
|
holocytochrome c synthase |
| chr7_+_2458833 | 0.08 |
ENSRNOT00000085092
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr2_+_193572053 | 0.08 |
ENSRNOT00000056479
|
AABR07012329.1
|
|
| chr3_-_161027943 | 0.08 |
ENSRNOT00000078633
|
Spint3
|
serine peptidase inhibitor, Kunitz type, 3 |
| chr1_-_201110928 | 0.08 |
ENSRNOT00000027721
|
Nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr15_+_48327461 | 0.08 |
ENSRNOT00000018071
|
Ints9
|
integrator complex subunit 9 |
| chrX_-_111942749 | 0.08 |
ENSRNOT00000087583
|
AABR07040840.1
|
|
| chr1_-_216750844 | 0.08 |
ENSRNOT00000054862
|
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr8_+_50604768 | 0.08 |
ENSRNOT00000059383
|
AC135409.1
|
|
| chr4_-_157252565 | 0.08 |
ENSRNOT00000079947
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr7_+_3320103 | 0.08 |
ENSRNOT00000090381
|
Cd63
|
Cd63 molecule |
| chr3_+_140106766 | 0.08 |
ENSRNOT00000014046
|
Naa20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
| chr1_-_263920934 | 0.08 |
ENSRNOT00000090755
ENSRNOT00000017006 |
Bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr1_+_65537640 | 0.08 |
ENSRNOT00000031007
|
Ube2m
|
ubiquitin-conjugating enzyme E2M |
| chr1_+_78833157 | 0.08 |
ENSRNOT00000022461
|
Ptgir
|
prostaglandin I2 (prostacyclin) receptor (IP) |
| chr7_+_11301957 | 0.08 |
ENSRNOT00000027847
|
Tjp3
|
tight junction protein 3 |
| chr1_+_198259496 | 0.08 |
ENSRNOT00000026955
|
Fam57b
|
family with sequence similarity 57, member B |
| chr10_-_13542077 | 0.08 |
ENSRNOT00000008736
|
Atp6v0c
|
ATPase H+ transporting V0 subunit C |
| chr5_-_152464850 | 0.08 |
ENSRNOT00000021937
|
Zfp593
|
zinc finger protein 593 |
| chr4_-_147719072 | 0.08 |
ENSRNOT00000014493
|
Rpl32
|
ribosomal protein L32 |
| chr16_+_54332660 | 0.08 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr3_+_168124673 | 0.08 |
ENSRNOT00000070809
|
Pfdn4
|
prefoldin subunit 4 |
| chr7_+_11784341 | 0.08 |
ENSRNOT00000026155
|
Plekhj1
|
pleckstrin homology domain containing J1 |
| chr12_-_16126953 | 0.07 |
ENSRNOT00000001682
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr4_-_119714302 | 0.07 |
ENSRNOT00000013299
|
Rab43
|
RAB43, member RAS oncogene family |
| chr3_-_168018410 | 0.07 |
ENSRNOT00000087579
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr1_+_225037737 | 0.07 |
ENSRNOT00000077959
|
Bscl2
|
BSCL2, seipin lipid droplet biogenesis associated |
| chr2_+_188784222 | 0.07 |
ENSRNOT00000028095
|
Pmvk
|
phosphomevalonate kinase |
| chrX_-_14220662 | 0.07 |
ENSRNOT00000045753
|
Srpx
|
sushi-repeat-containing protein, X-linked |
| chr7_+_125576327 | 0.07 |
ENSRNOT00000016679
|
Prr5
|
proline rich 5 |
| chr6_+_108488678 | 0.07 |
ENSRNOT00000016089
|
Isca2
|
iron-sulfur cluster assembly 2 |
| chr13_-_36022197 | 0.07 |
ENSRNOT00000091280
|
Cfap221
|
cilia and flagella associated protein 221 |
| chr20_+_13827132 | 0.07 |
ENSRNOT00000001664
|
Ddt
|
D-dopachrome tautomerase |
| chr9_-_82673898 | 0.07 |
ENSRNOT00000027165
|
Chpf
|
chondroitin polymerizing factor |
| chr1_-_77830399 | 0.07 |
ENSRNOT00000052231
|
LOC100360449
|
ribosomal protein L9-like |
| chr10_+_11046221 | 0.07 |
ENSRNOT00000005109
|
Nmral1
|
NmrA like redox sensor 1 |
| chr20_+_3990820 | 0.07 |
ENSRNOT00000000528
|
Psmb8
|
proteasome subunit beta 8 |
| chr8_-_91338843 | 0.07 |
ENSRNOT00000012999
|
Elovl4
|
ELOVL fatty acid elongase 4 |
| chr7_+_2458264 | 0.07 |
ENSRNOT00000003550
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr1_+_191829555 | 0.07 |
ENSRNOT00000067138
|
Scnn1b
|
sodium channel epithelial 1 beta subunit |
| chr1_+_216254979 | 0.07 |
ENSRNOT00000046669
|
Tssc4
|
tumor suppressing subtransferable candidate 4 |
| chrX_-_13116743 | 0.07 |
ENSRNOT00000004305
|
Mid1ip1
|
MID1 interacting protein 1 |
| chr10_+_43601689 | 0.07 |
ENSRNOT00000029238
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr14_-_37475418 | 0.07 |
ENSRNOT00000002996
|
Ociad1
|
OCIA domain containing 1 |
| chr6_-_108488330 | 0.07 |
ENSRNOT00000016076
|
Npc2
|
NPC intracellular cholesterol transporter 2 |
| chr7_+_34326087 | 0.07 |
ENSRNOT00000006971
|
Hal
|
histidine ammonia lyase |
| chr1_-_1474846 | 0.07 |
ENSRNOT00000028992
|
Raet1d
|
retinoic acid early transcript delta |
| chr7_-_18585313 | 0.07 |
ENSRNOT00000010399
|
March2
|
membrane associated ring-CH-type finger 2 |
| chr5_+_131719922 | 0.07 |
ENSRNOT00000010524
|
Spata6
|
spermatogenesis associated 6 |
| chr9_+_102862890 | 0.07 |
ENSRNOT00000050494
ENSRNOT00000080129 |
Fam174a
|
family with sequence similarity 174, member A |
| chr5_-_139505065 | 0.07 |
ENSRNOT00000013899
|
Ctps1
|
CTP synthase 1 |
| chr1_+_167538744 | 0.07 |
ENSRNOT00000093070
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr17_+_76079720 | 0.07 |
ENSRNOT00000073933
|
Proser2
|
proline and serine rich 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp524
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.2 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 0.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.3 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.2 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 0.0 | 0.3 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.1 | GO:0090648 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) response to environmental enrichment(GO:0090648) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:1903769 | negative regulation of cell proliferation in bone marrow(GO:1903769) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.1 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0002876 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.0 | 0.2 | GO:0015746 | citrate transport(GO:0015746) |
| 0.0 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:1903195 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) TRAIL receptor biosynthetic process(GO:0045557) regulation of TRAIL receptor biosynthetic process(GO:0045560) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.0 | 0.2 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0035419 | detection of peptidoglycan(GO:0032499) activation of MAPK activity involved in innate immune response(GO:0035419) |
| 0.0 | 0.1 | GO:0061010 | inner cell mass cellular morphogenesis(GO:0001828) endodermal cell fate determination(GO:0007493) gall bladder development(GO:0061010) |
| 0.0 | 0.3 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 0.4 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.1 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) |
| 0.0 | 0.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.0 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.1 | GO:0014028 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) notochord formation(GO:0014028) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0000053 | argininosuccinate metabolic process(GO:0000053) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.0 | GO:1904313 | response to methamphetamine hydrochloride(GO:1904313) |
| 0.0 | 0.1 | GO:0033123 | positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.0 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.0 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.0 | GO:0016128 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.0 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.0 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.0 | 0.0 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.0 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.0 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 0.1 | GO:0035826 | rubidium ion transport(GO:0035826) |
| 0.0 | 0.1 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.0 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.0 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:1990622 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.0 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.2 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.0 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.0 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.4 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.1 | 0.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.2 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) |
| 0.1 | 0.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0034235 | GPI-anchor transamidase activity(GO:0003923) GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.0 | GO:0051903 | all-trans retinal binding(GO:0005503) benzaldehyde dehydrogenase activity(GO:0019115) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0050700 | peptidoglycan binding(GO:0042834) CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |