Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
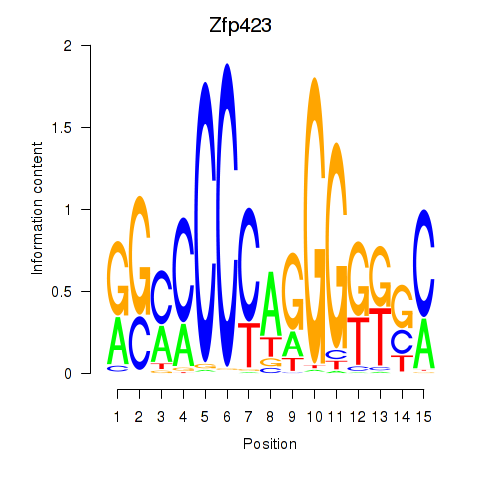
Results for Zfp423
Z-value: 0.30
Transcription factors associated with Zfp423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp423
|
ENSRNOG00000014658 | zinc finger protein 423 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp423 | rn6_v1_chr19_+_20147037_20147037 | 0.94 | 1.6e-02 | Click! |
Activity profile of Zfp423 motif
Sorted Z-values of Zfp423 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_77945016 | 0.19 |
ENSRNOT00000076223
|
Zfp37
|
zinc finger protein 37 |
| chr10_-_18131562 | 0.12 |
ENSRNOT00000006676
|
Tlx3
|
T-cell leukemia, homeobox 3 |
| chr1_+_266953139 | 0.11 |
ENSRNOT00000054696
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr4_+_142453013 | 0.11 |
ENSRNOT00000056573
|
AABR07061755.1
|
|
| chr12_-_47793534 | 0.10 |
ENSRNOT00000001588
|
Fam222a
|
family with sequence similarity 222, member A |
| chr1_-_198316882 | 0.09 |
ENSRNOT00000085304
ENSRNOT00000064985 |
Taok2
|
TAO kinase 2 |
| chr7_+_130532435 | 0.08 |
ENSRNOT00000092672
ENSRNOT00000092535 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr1_-_259484569 | 0.08 |
ENSRNOT00000021289
ENSRNOT00000021114 ENSRNOT00000021229 ENSRNOT00000021347 ENSRNOT00000021226 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr19_+_55094585 | 0.07 |
ENSRNOT00000068452
|
Zfpm1
|
zinc finger protein, multitype 1 |
| chr7_+_120067379 | 0.06 |
ENSRNOT00000011270
|
Cdc42ep1
|
CDC42 effector protein 1 |
| chr15_-_28733513 | 0.06 |
ENSRNOT00000078180
|
Sall2
|
spalt-like transcription factor 2 |
| chr4_-_156427755 | 0.06 |
ENSRNOT00000085675
ENSRNOT00000014030 |
LOC103690024
|
peroxisomal targeting signal 1 receptor-like |
| chr4_-_157008947 | 0.06 |
ENSRNOT00000073341
|
Pex5
|
peroxisomal biogenesis factor 5 |
| chr1_+_198932870 | 0.06 |
ENSRNOT00000055003
|
Fbrs
|
fibrosin |
| chr13_-_90710148 | 0.06 |
ENSRNOT00000010113
ENSRNOT00000080638 |
Kcnj9
|
potassium voltage-gated channel subfamily J member 9 |
| chr1_-_226924244 | 0.05 |
ENSRNOT00000028971
|
Tmem132a
|
transmembrane protein 132A |
| chr1_-_225948626 | 0.05 |
ENSRNOT00000077823
|
Incenp
|
inner centromere protein |
| chr1_-_226152524 | 0.05 |
ENSRNOT00000027756
|
Fads2
|
fatty acid desaturase 2 |
| chr1_+_40529045 | 0.04 |
ENSRNOT00000026564
|
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr13_+_51384562 | 0.04 |
ENSRNOT00000077928
|
Kdm5b
|
lysine demethylase 5B |
| chr15_-_52210746 | 0.04 |
ENSRNOT00000046054
|
Bmp1
|
bone morphogenetic protein 1 |
| chr7_-_144960527 | 0.03 |
ENSRNOT00000086554
|
Zfp385a
|
zinc finger protein 385A |
| chr19_-_55490426 | 0.03 |
ENSRNOT00000081800
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr7_-_14189688 | 0.03 |
ENSRNOT00000037456
|
Notch3
|
notch 3 |
| chr16_-_74264142 | 0.03 |
ENSRNOT00000026067
|
Dkk4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr1_-_131454689 | 0.03 |
ENSRNOT00000014152
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr12_-_17358617 | 0.02 |
ENSRNOT00000001733
|
Gpr146
|
G protein-coupled receptor 146 |
| chr13_+_35554964 | 0.02 |
ENSRNOT00000072632
|
Tmem185b
|
transmembrane protein 185B |
| chr7_-_3074359 | 0.02 |
ENSRNOT00000019738
|
Ikzf4
|
IKAROS family zinc finger 4 |
| chr2_-_210413985 | 0.02 |
ENSRNOT00000025036
|
Strip1
|
striatin interacting protein 1 |
| chr3_+_56868167 | 0.02 |
ENSRNOT00000087134
|
Gad1
|
glutamate decarboxylase 1 |
| chrX_+_140878216 | 0.02 |
ENSRNOT00000075840
ENSRNOT00000077026 |
Zic3
|
Zic family member 3 |
| chr6_-_44361908 | 0.02 |
ENSRNOT00000009491
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr9_+_53120656 | 0.02 |
ENSRNOT00000043714
|
Pms1
|
PMS1 homolog 1, mismatch repair system component |
| chr1_-_101856881 | 0.01 |
ENSRNOT00000028615
|
Grin2d
|
glutamate ionotropic receptor NMDA type subunit 2D |
| chr7_-_70926903 | 0.01 |
ENSRNOT00000031005
|
Lrp1
|
LDL receptor related protein 1 |
| chr8_+_82038967 | 0.01 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr10_+_80790168 | 0.01 |
ENSRNOT00000073315
ENSRNOT00000075163 |
Car10
|
carbonic anhydrase 10 |
| chr1_-_170464341 | 0.01 |
ENSRNOT00000024850
|
Trim3
|
tripartite motif-containing 3 |
| chr16_+_47665816 | 0.01 |
ENSRNOT00000030519
|
Cdkn2aip
|
CDKN2A interacting protein |
| chrX_-_72515320 | 0.01 |
ENSRNOT00000076725
|
Phka1
|
phosphorylase kinase, alpha 1 |
| chr3_-_122920684 | 0.01 |
ENSRNOT00000028818
|
Cpxm1
|
carboxypeptidase X (M14 family), member 1 |
| chr1_-_82546937 | 0.01 |
ENSRNOT00000028100
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr2_-_61949926 | 0.01 |
ENSRNOT00000025966
|
Npr3
|
natriuretic peptide receptor 3 |
| chr1_+_226091774 | 0.01 |
ENSRNOT00000027693
|
Fads3
|
fatty acid desaturase 3 |
| chr10_-_104097812 | 0.01 |
ENSRNOT00000004867
|
Sumo2
|
small ubiquitin-like modifier 2 |
| chr9_+_117538346 | 0.01 |
ENSRNOT00000022849
|
Epb41l3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr4_-_157009674 | 0.00 |
ENSRNOT00000071625
|
Pex5
|
peroxisomal biogenesis factor 5 |
| chr7_-_11444786 | 0.00 |
ENSRNOT00000027323
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr19_-_11057254 | 0.00 |
ENSRNOT00000025559
|
Herpud1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr5_+_138279506 | 0.00 |
ENSRNOT00000084651
|
P3h1
|
prolyl 3-hydroxylase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp423
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0090202 | mitral valve formation(GO:0003192) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:1901090 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.1 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.1 | GO:1904008 | response to monosodium glutamate(GO:1904008) |
| 0.0 | 0.0 | GO:0015942 | formate metabolic process(GO:0015942) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |