Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
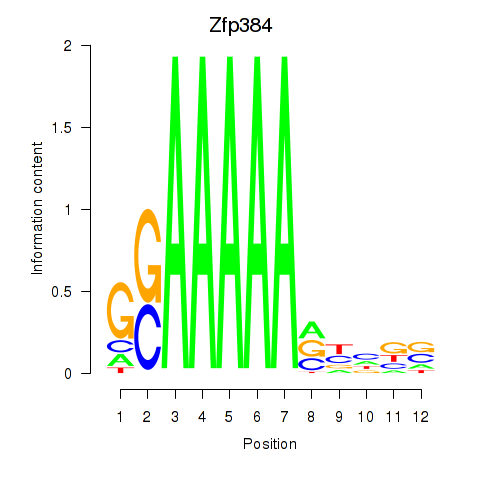
Results for Zfp384
Z-value: 0.88
Transcription factors associated with Zfp384
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp384
|
ENSRNOG00000017066 | zinc finger protein 384 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp384 | rn6_v1_chr4_+_157538303_157538303 | -0.61 | 2.8e-01 | Click! |
Activity profile of Zfp384 motif
Sorted Z-values of Zfp384 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_24242642 | 0.43 |
ENSRNOT00000024323
|
Rnf182
|
ring finger protein 182 |
| chr15_-_23969011 | 0.32 |
ENSRNOT00000014821
|
Gch1
|
GTP cyclohydrolase 1 |
| chr1_+_218466289 | 0.32 |
ENSRNOT00000017948
|
Mrgprf
|
MAS related GPR family member F |
| chr4_+_438668 | 0.32 |
ENSRNOT00000008951
|
En2
|
engrailed homeobox 2 |
| chr14_-_91996774 | 0.29 |
ENSRNOT00000005851
|
Ddc
|
dopa decarboxylase |
| chr4_-_482645 | 0.27 |
ENSRNOT00000062073
ENSRNOT00000071713 |
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr15_-_34550850 | 0.27 |
ENSRNOT00000027794
ENSRNOT00000090228 |
Cbln3
|
cerebellin 3 precursor |
| chr12_-_38782010 | 0.27 |
ENSRNOT00000001813
|
Wdr66
|
WD repeat domain 66 |
| chr13_+_36532758 | 0.21 |
ENSRNOT00000083007
|
En1
|
engrailed 1 |
| chr18_-_5314511 | 0.20 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr3_+_152226070 | 0.20 |
ENSRNOT00000073201
|
AABR07054397.1
|
|
| chr4_+_94696965 | 0.20 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr17_-_43776460 | 0.19 |
ENSRNOT00000089055
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chrX_+_14890828 | 0.19 |
ENSRNOT00000080826
|
LOC102552805
|
uncharacterized LOC102552805 |
| chr15_+_30413488 | 0.19 |
ENSRNOT00000071781
|
AABR07017733.1
|
|
| chr2_-_211017778 | 0.18 |
ENSRNOT00000026883
|
Sypl2
|
synaptophysin-like 2 |
| chr14_+_87448692 | 0.18 |
ENSRNOT00000077177
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr15_+_32386816 | 0.18 |
ENSRNOT00000060322
|
AABR07017901.1
|
|
| chr17_-_44793927 | 0.18 |
ENSRNOT00000086309
|
Hist1h2bo
|
histone cluster 1 H2B family member o |
| chr8_+_2635851 | 0.17 |
ENSRNOT00000061973
|
Casp4
|
caspase 4 |
| chr6_-_123577695 | 0.17 |
ENSRNOT00000006604
|
Foxn3
|
forkhead box N3 |
| chr14_-_44375804 | 0.17 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr10_-_93679974 | 0.17 |
ENSRNOT00000009316
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr9_-_17212628 | 0.17 |
ENSRNOT00000002656
ENSRNOT00000007876 |
RGD1562399
|
similar to 40S ribosomal protein S2 |
| chr17_+_18031228 | 0.17 |
ENSRNOT00000081708
|
Tpmt
|
thiopurine S-methyltransferase |
| chr20_+_40618128 | 0.16 |
ENSRNOT00000001075
ENSRNOT00000057364 |
Pkib
|
cAMP-dependent protein kinase inhibitor beta |
| chr2_+_257295193 | 0.16 |
ENSRNOT00000052249
|
AABR07013798.1
|
|
| chr18_+_70974885 | 0.16 |
ENSRNOT00000025198
|
RGD1562987
|
similar to cDNA sequence BC031181 |
| chr4_-_170912629 | 0.16 |
ENSRNOT00000055691
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr1_-_88881460 | 0.15 |
ENSRNOT00000028287
|
Hcst
|
hematopoietic cell signal transducer |
| chr20_+_40618623 | 0.15 |
ENSRNOT00000082043
|
Pkib
|
cAMP-dependent protein kinase inhibitor beta |
| chr5_+_159606647 | 0.15 |
ENSRNOT00000051693
|
Rps21-ps1
|
ribosomal protein S21, pseudogene 1 |
| chr5_+_151181559 | 0.15 |
ENSRNOT00000085674
|
Fgr
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr19_-_43911057 | 0.15 |
ENSRNOT00000026017
|
Ctrb1
|
chymotrypsinogen B1 |
| chr1_+_88620317 | 0.14 |
ENSRNOT00000075116
|
Cox7a1
|
cytochrome c oxidase subunit 7A1 |
| chr6_-_102095201 | 0.14 |
ENSRNOT00000014125
|
Plek2
|
pleckstrin 2 |
| chr17_+_44763598 | 0.14 |
ENSRNOT00000079880
|
Hist1h3b
|
histone cluster 1, H3b |
| chr8_-_40883880 | 0.14 |
ENSRNOT00000075593
|
LOC102550797
|
disks large homolog 5-like |
| chr8_-_132027006 | 0.14 |
ENSRNOT00000050963
|
LOC367195
|
similar to 60S RIBOSOMAL PROTEIN L7 |
| chr5_+_144106802 | 0.14 |
ENSRNOT00000035637
ENSRNOT00000079779 |
Lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr17_-_32015071 | 0.14 |
ENSRNOT00000070876
|
LOC100911107
|
leukocyte elastase inhibitor A-like |
| chr17_+_44760056 | 0.14 |
ENSRNOT00000052073
|
LOC690171
|
similar to H3 histone, family 3B |
| chr5_-_135498822 | 0.14 |
ENSRNOT00000023072
|
Akr1a1
|
aldo-keto reductase family 1 member A1 |
| chr2_-_1511591 | 0.13 |
ENSRNOT00000087906
ENSRNOT00000014709 |
Cast
|
calpastatin |
| chr11_-_71601662 | 0.13 |
ENSRNOT00000002397
|
Tctex1d2
|
Tctex1 domain containing 2 |
| chr17_+_72429618 | 0.13 |
ENSRNOT00000026187
|
Gata3
|
GATA binding protein 3 |
| chr12_-_2661565 | 0.13 |
ENSRNOT00000064859
|
Cd209f
|
CD209f antigen |
| chr17_+_44794130 | 0.13 |
ENSRNOT00000077571
|
Hist1h2ac
|
histone cluster 1, H2ac |
| chr15_-_61873406 | 0.13 |
ENSRNOT00000045115
|
LOC100360244
|
LRRGT00053-like |
| chr4_+_172119331 | 0.13 |
ENSRNOT00000010579
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr15_-_64191115 | 0.13 |
ENSRNOT00000013868
|
LOC100361180
|
40S ribosomal protein S17-like |
| chr9_+_95274707 | 0.13 |
ENSRNOT00000045163
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr20_+_1889652 | 0.13 |
ENSRNOT00000068508
ENSRNOT00000000996 |
Olr1746
|
olfactory receptor 1746 |
| chr11_-_72109964 | 0.13 |
ENSRNOT00000058917
|
AABR07034445.1
|
|
| chr9_-_121725716 | 0.13 |
ENSRNOT00000087405
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr2_+_195603599 | 0.13 |
ENSRNOT00000028263
|
C2cd4d
|
C2 calcium-dependent domain containing 4D |
| chr3_-_24601063 | 0.13 |
ENSRNOT00000037043
|
LOC100362366
|
40S ribosomal protein S17-like |
| chr16_-_19988004 | 0.12 |
ENSRNOT00000024424
|
Tmem221
|
transmembrane protein 221 |
| chr3_+_151691515 | 0.12 |
ENSRNOT00000052236
|
AC118414.1
|
|
| chr19_+_14523554 | 0.12 |
ENSRNOT00000084271
ENSRNOT00000064731 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr1_-_190598122 | 0.12 |
ENSRNOT00000044384
|
Pdzd9
|
PDZ domain containing 9 |
| chr3_-_48451650 | 0.12 |
ENSRNOT00000007356
|
Gcg
|
glucagon |
| chr10_+_74724549 | 0.12 |
ENSRNOT00000009077
|
Tex14
|
testis expressed 14, intercellular bridge forming factor |
| chr13_-_111474411 | 0.12 |
ENSRNOT00000072729
|
Hhat
|
hedgehog acyltransferase |
| chr19_+_37873515 | 0.12 |
ENSRNOT00000026163
ENSRNOT00000089921 |
Pskh1
|
protein serine kinase H1 |
| chr12_+_11191812 | 0.12 |
ENSRNOT00000033537
|
Atp5j2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2 |
| chr13_-_45318822 | 0.12 |
ENSRNOT00000091514
ENSRNOT00000005143 |
Cxcr4
|
C-X-C motif chemokine receptor 4 |
| chr2_+_24546536 | 0.12 |
ENSRNOT00000014036
|
Otp
|
orthopedia homeobox |
| chr2_-_198458041 | 0.12 |
ENSRNOT00000082450
|
Fcgr1a
|
Fc fragment of IgG receptor Ia |
| chr19_+_10596960 | 0.11 |
ENSRNOT00000021769
|
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr5_-_128446725 | 0.11 |
ENSRNOT00000011272
|
Btf3l4
|
basic transcription factor 3-like 4 |
| chr12_-_21832813 | 0.11 |
ENSRNOT00000075280
|
Cldn3
|
claudin 3 |
| chr3_-_59166356 | 0.11 |
ENSRNOT00000047713
|
AABR07052519.1
|
|
| chr4_+_119680112 | 0.11 |
ENSRNOT00000013282
|
Gp9
|
glycoprotein IX (platelet) |
| chr4_+_149877889 | 0.11 |
ENSRNOT00000038521
|
Zfp637
|
zinc finger protein 637 |
| chr1_+_207508414 | 0.11 |
ENSRNOT00000054896
|
Nps
|
neuropeptide S |
| chr12_+_19196611 | 0.11 |
ENSRNOT00000001801
|
Azgp1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr7_+_122160171 | 0.11 |
ENSRNOT00000074499
|
LOC100362980
|
CG3918-like |
| chr5_-_134927235 | 0.11 |
ENSRNOT00000016751
|
Uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chr17_-_43675934 | 0.11 |
ENSRNOT00000081345
|
Hist1h1t
|
histone cluster 1 H1 family member t |
| chr1_-_37990007 | 0.11 |
ENSRNOT00000081054
|
AABR07001100.1
|
|
| chr10_+_71202456 | 0.11 |
ENSRNOT00000076893
|
Hnf1b
|
HNF1 homeobox B |
| chr17_-_2705123 | 0.11 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr2_+_243550627 | 0.11 |
ENSRNOT00000085067
ENSRNOT00000083682 |
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr4_-_116278615 | 0.11 |
ENSRNOT00000020505
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chrX_-_77418525 | 0.11 |
ENSRNOT00000092303
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr7_-_186360 | 0.11 |
ENSRNOT00000073828
|
LOC100912293
|
uncharacterized LOC100912293 |
| chr1_+_264504591 | 0.11 |
ENSRNOT00000050076
|
Pax2
|
paired box 2 |
| chr8_+_48727618 | 0.11 |
ENSRNOT00000029546
|
Rps25
|
ribosomal protein s25 |
| chr1_+_42170281 | 0.11 |
ENSRNOT00000092879
|
Vip
|
vasoactive intestinal peptide |
| chr8_-_133002201 | 0.11 |
ENSRNOT00000008772
|
Ccr1
|
C-C motif chemokine receptor 1 |
| chr10_+_55712043 | 0.10 |
ENSRNOT00000010141
|
Aloxe3
|
arachidonate lipoxygenase 3 |
| chr8_+_115151627 | 0.10 |
ENSRNOT00000064252
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr6_-_102272777 | 0.10 |
ENSRNOT00000014775
|
Pigh
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr3_-_113423149 | 0.10 |
ENSRNOT00000021008
|
Serinc4
|
serine incorporator 4 |
| chr19_-_645937 | 0.10 |
ENSRNOT00000016521
|
Car7
|
carbonic anhydrase 7 |
| chr10_-_83898527 | 0.10 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr11_+_36555416 | 0.10 |
ENSRNOT00000064981
|
Sh3bgr
|
SH3 domain binding glutamate-rich protein |
| chr7_+_117030289 | 0.10 |
ENSRNOT00000081286
|
Zfp707
|
zinc finger protein 707 |
| chr7_+_2435553 | 0.10 |
ENSRNOT00000065593
|
Prim1
|
primase (DNA) subunit 1 |
| chr10_+_40247436 | 0.10 |
ENSRNOT00000079830
|
Gpx3
|
glutathione peroxidase 3 |
| chr18_-_36513317 | 0.10 |
ENSRNOT00000025453
|
Plac8l1
|
PLAC8-like 1 |
| chr5_+_168078748 | 0.10 |
ENSRNOT00000024798
|
Uts2
|
urotensin 2 |
| chr18_-_63394690 | 0.10 |
ENSRNOT00000090078
ENSRNOT00000029431 |
Cep76
|
centrosomal protein 76 |
| chr17_+_15852548 | 0.10 |
ENSRNOT00000022203
|
Card19
|
caspase recruitment domain family, member 19 |
| chrX_+_1787266 | 0.10 |
ENSRNOT00000011183
|
Ndufb11
|
NADH:ubiquinone oxidoreductase subunit B11 |
| chr19_+_53044379 | 0.10 |
ENSRNOT00000072369
|
Foxc2
|
forkhead box C2 |
| chr15_-_44442875 | 0.10 |
ENSRNOT00000017939
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr12_+_46042413 | 0.10 |
ENSRNOT00000046882
|
Ccdc60
|
coiled-coil domain containing 60 |
| chr2_-_208215187 | 0.10 |
ENSRNOT00000047836
|
Rap1a
|
RAP1A, member of RAS oncogene family |
| chr6_+_86131242 | 0.10 |
ENSRNOT00000039337
|
Fau
|
FAU, ubiquitin like and ribosomal protein S30 fusion |
| chr5_-_38923095 | 0.10 |
ENSRNOT00000009146
|
AABR07047593.1
|
|
| chr17_+_29638677 | 0.10 |
ENSRNOT00000068617
|
Rpp40
|
ribonuclease P/MRP 40 subunit |
| chr18_-_7081356 | 0.10 |
ENSRNOT00000021253
|
Chst9
|
carbohydrate sulfotransferase 9 |
| chr4_-_66624912 | 0.10 |
ENSRNOT00000064891
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr1_-_225830300 | 0.10 |
ENSRNOT00000072584
ENSRNOT00000027502 |
Scgb2a2
|
secretoglobin, family 2A, member 2 |
| chr5_+_70441123 | 0.10 |
ENSRNOT00000087517
|
Fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr7_+_79638046 | 0.09 |
ENSRNOT00000029419
|
Zfpm2
|
zinc finger protein, multitype 2 |
| chr1_+_89008117 | 0.09 |
ENSRNOT00000028401
|
Hspb6
|
heat shock protein family B (small) member 6 |
| chr1_+_234363994 | 0.09 |
ENSRNOT00000018137
|
Rorb
|
RAR-related orphan receptor B |
| chr8_-_49227273 | 0.09 |
ENSRNOT00000050878
|
Atp5l
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G |
| chr8_+_98755104 | 0.09 |
ENSRNOT00000056562
|
Zic4
|
Zic family member 4 |
| chr16_+_54386513 | 0.09 |
ENSRNOT00000080696
|
AABR07025896.1
|
|
| chr10_+_69423086 | 0.09 |
ENSRNOT00000000256
|
Ccl7
|
C-C motif chemokine ligand 7 |
| chr1_-_164441167 | 0.09 |
ENSRNOT00000023935
|
Rps3
|
ribosomal protein S3 |
| chr8_+_48438259 | 0.09 |
ENSRNOT00000059813
|
Mfrp
|
membrane frizzled-related protein |
| chr2_+_195940715 | 0.09 |
ENSRNOT00000042985
|
LOC100363268
|
rCG31129-like |
| chr19_-_10212050 | 0.09 |
ENSRNOT00000082822
|
Tepp
|
testis, prostate and placenta expressed |
| chr3_-_103745236 | 0.09 |
ENSRNOT00000006876
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr7_-_13108630 | 0.09 |
ENSRNOT00000087038
ENSRNOT00000080491 |
Giot1
|
gonadotropin inducible ovarian transcription factor 1 |
| chr1_-_55895636 | 0.09 |
ENSRNOT00000029537
|
Vom2r9
|
vomeronasal 2 receptor, 9 |
| chr1_-_276012351 | 0.09 |
ENSRNOT00000045642
|
AABR07007000.1
|
|
| chr15_+_33333420 | 0.09 |
ENSRNOT00000049927
|
RGD1308430
|
similar to 1700123O20Rik protein |
| chr8_-_115145810 | 0.09 |
ENSRNOT00000079470
|
Abhd14a
|
abhydrolase domain containing 14A |
| chr6_+_128738388 | 0.09 |
ENSRNOT00000050204
|
Rpl6-ps1
|
ribosomal protein L6, pseudogene 1 |
| chrX_+_20520034 | 0.09 |
ENSRNOT00000093170
|
FAM120C
|
family with sequence similarity 120C |
| chr7_+_12314848 | 0.09 |
ENSRNOT00000028969
|
Gamt
|
guanidinoacetate N-methyltransferase |
| chr7_-_30397930 | 0.09 |
ENSRNOT00000010414
|
Actr6
|
ARP6 actin-related protein 6 homolog |
| chr8_-_58647933 | 0.09 |
ENSRNOT00000038830
|
Tnfaip8l3
|
TNF alpha induced protein 8 like 3 |
| chr14_-_15008119 | 0.09 |
ENSRNOT00000002834
ENSRNOT00000084617 |
Mrpl1
|
mitochondrial ribosomal protein L1 |
| chr13_-_37492680 | 0.09 |
ENSRNOT00000003441
|
Htr5b
|
5-hydroxytryptamine (serotonin) receptor 5B |
| chr14_-_77279831 | 0.09 |
ENSRNOT00000007115
|
Tmem128
|
transmembrane protein 128 |
| chr6_+_93385457 | 0.09 |
ENSRNOT00000010300
|
Actr10
|
actin-related protein 10 homolog |
| chr14_-_72970329 | 0.09 |
ENSRNOT00000006325
|
Pp2d1
|
protein phosphatase 2C-like domain containing 1 |
| chr4_+_147844658 | 0.09 |
ENSRNOT00000042673
|
H1foo
|
H1 histone family, member O, oocyte-specific |
| chr9_-_65957101 | 0.09 |
ENSRNOT00000014094
|
Mpp4
|
membrane palmitoylated protein 4 |
| chr1_+_257970345 | 0.09 |
ENSRNOT00000088853
|
Cyp2c11
|
cytochrome P450, subfamily 2, polypeptide 11 |
| chr6_-_127337791 | 0.09 |
ENSRNOT00000032015
|
Ifi27l2b
|
interferon, alpha-inducible protein 27 like 2B |
| chr3_+_151335292 | 0.08 |
ENSRNOT00000073642
|
Mmp24
|
matrix metallopeptidase 24 |
| chr3_-_2490392 | 0.08 |
ENSRNOT00000014993
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr5_-_167999853 | 0.08 |
ENSRNOT00000087402
|
Park7
|
Parkinsonism associated deglycase |
| chr18_+_35574002 | 0.08 |
ENSRNOT00000093442
ENSRNOT00000070817 ENSRNOT00000093356 |
Myot
|
myotilin |
| chr9_-_17693200 | 0.08 |
ENSRNOT00000026716
|
Mrpl14
|
mitochondrial ribosomal protein L14 |
| chr16_-_81693000 | 0.08 |
ENSRNOT00000092353
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr14_+_100415668 | 0.08 |
ENSRNOT00000008057
|
C1d
|
C1D nuclear receptor co-repressor |
| chr5_+_141560192 | 0.08 |
ENSRNOT00000023354
|
Mycbp
|
Myc binding protein |
| chr16_+_81616604 | 0.08 |
ENSRNOT00000026392
ENSRNOT00000057740 |
Adprhl1
Grtp1
|
ADP-ribosylhydrolase like 1 growth hormone regulated TBC protein 1 |
| chr7_+_35125424 | 0.08 |
ENSRNOT00000085978
ENSRNOT00000010117 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr7_-_106653567 | 0.08 |
ENSRNOT00000042522
|
Oc90
|
otoconin 90 |
| chr7_-_98960736 | 0.08 |
ENSRNOT00000011984
ENSRNOT00000080155 |
Mtss1
|
MTSS1, I-BAR domain containing |
| chr13_-_80703615 | 0.08 |
ENSRNOT00000004599
|
Fmo4
|
flavin containing monooxygenase 4 |
| chr8_-_63291966 | 0.08 |
ENSRNOT00000077762
|
AABR07070278.1
|
|
| chr10_+_71217966 | 0.08 |
ENSRNOT00000076192
|
Hnf1b
|
HNF1 homeobox B |
| chr14_-_43143973 | 0.08 |
ENSRNOT00000003248
|
Uchl1
|
ubiquitin C-terminal hydrolase L1 |
| chr8_+_96395317 | 0.08 |
ENSRNOT00000044521
|
Trim43a
|
tripartite motif-containing 43A |
| chr15_+_47455690 | 0.08 |
ENSRNOT00000075276
|
LOC100910401
|
serine protease 55-like |
| chr10_-_59743037 | 0.08 |
ENSRNOT00000026168
|
Emc6
|
ER membrane protein complex subunit 6 |
| chr6_+_127327959 | 0.08 |
ENSRNOT00000012296
|
Ifi27
|
interferon, alpha-inducible protein 27 |
| chr17_-_53102342 | 0.08 |
ENSRNOT00000073834
|
Psma2
|
proteasome subunit alpha 2 |
| chr1_+_248132090 | 0.08 |
ENSRNOT00000022056
|
Il33
|
interleukin 33 |
| chr9_+_20213588 | 0.08 |
ENSRNOT00000089341
|
LOC100911515
|
triosephosphate isomerase-like |
| chr20_-_3372070 | 0.08 |
ENSRNOT00000080770
|
Dhx16
|
DEAH-box helicase 16 |
| chr14_-_10446909 | 0.08 |
ENSRNOT00000002962
|
Mrps18c
|
mitochondrial ribosomal protein S18C |
| chr14_-_18745457 | 0.08 |
ENSRNOT00000003778
|
Cxcl1
|
C-X-C motif chemokine ligand 1 |
| chr9_-_65442257 | 0.08 |
ENSRNOT00000037660
|
Fam126b
|
family with sequence similarity 126, member B |
| chr1_-_48360131 | 0.08 |
ENSRNOT00000051927
ENSRNOT00000023116 |
Slc22a2
|
solute carrier family 22 member 2 |
| chr7_+_27221152 | 0.08 |
ENSRNOT00000031297
|
LOC691921
|
hypothetical protein LOC691921 |
| chr17_+_24654902 | 0.08 |
ENSRNOT00000041960
|
Tfap2a
|
transcription factor AP-2 alpha |
| chr4_+_84423653 | 0.08 |
ENSRNOT00000012655
|
Chn2
|
chimerin 2 |
| chr16_-_11214948 | 0.08 |
ENSRNOT00000023369
|
LOC690468
|
similar to 60S ribosomal protein L38 |
| chr8_+_76426335 | 0.08 |
ENSRNOT00000085496
|
Gtf2a2
|
general transcription factor2A subunit 2 |
| chr9_+_37727942 | 0.08 |
ENSRNOT00000016511
ENSRNOT00000074276 |
LOC100912306
|
myotilin-like |
| chr10_+_31532250 | 0.08 |
ENSRNOT00000009238
|
Med7
|
mediator complex subunit 7 |
| chr10_-_59743315 | 0.08 |
ENSRNOT00000093646
|
Emc6
|
ER membrane protein complex subunit 6 |
| chr1_-_221321459 | 0.08 |
ENSRNOT00000028406
ENSRNOT00000084800 |
Pola2
|
DNA polymerase alpha 2, accessory subunit |
| chr9_+_46086984 | 0.08 |
ENSRNOT00000033733
|
Rpl31
|
ribosomal protein L31 |
| chrX_-_15347591 | 0.08 |
ENSRNOT00000037066
|
Timm17b
|
translocase of inner mitochondrial membrane 17b |
| chr4_+_100407658 | 0.08 |
ENSRNOT00000018562
|
Capg
|
capping actin protein, gelsolin like |
| chr8_-_61836796 | 0.08 |
ENSRNOT00000064903
ENSRNOT00000084009 |
Commd4
|
COMM domain containing 4 |
| chr3_-_113405829 | 0.08 |
ENSRNOT00000036823
|
Ell3
|
elongation factor for RNA polymerase II 3 |
| chr1_+_42170583 | 0.08 |
ENSRNOT00000093104
|
Vip
|
vasoactive intestinal peptide |
| chr2_+_248398917 | 0.08 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr6_-_47904163 | 0.08 |
ENSRNOT00000011333
|
Rps7
|
ribosomal protein S7 |
| chr1_+_64480929 | 0.08 |
ENSRNOT00000087016
|
Olr1l
|
olfactory receptor 1-like |
| chr3_+_160908769 | 0.08 |
ENSRNOT00000030054
|
Sys1
|
Sys1 golgi trafficking protein |
| chr2_+_209661244 | 0.08 |
ENSRNOT00000091973
|
AABR07012826.1
|
|
| chr2_+_120504130 | 0.08 |
ENSRNOT00000071944
|
Dnajc19
|
DnaJ heat shock protein family (Hsp40) member C19 |
| chr1_-_72632185 | 0.08 |
ENSRNOT00000066383
|
Tmem190
|
transmembrane protein 190 |
| chr1_+_101178104 | 0.08 |
ENSRNOT00000028072
|
Pth2
|
parathyroid hormone 2 |
| chr10_-_45923829 | 0.08 |
ENSRNOT00000035285
|
Olr1462
|
olfactory receptor 1462 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp384
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.1 | 0.3 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.1 | 0.2 | GO:0039020 | regulation of pronephros size(GO:0035565) pronephric nephron tubule development(GO:0039020) hepatoblast differentiation(GO:0061017) |
| 0.1 | 0.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 0.2 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.0 | 0.2 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:1903184 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) TRAIL receptor biosynthetic process(GO:0045557) regulation of TRAIL receptor biosynthetic process(GO:0045560) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.0 | 0.1 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.2 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.1 | GO:0070268 | cornification(GO:0070268) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:1903576 | response to L-arginine(GO:1903576) |
| 0.0 | 0.1 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.0 | 0.1 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.1 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.1 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.3 | GO:0019585 | uronic acid metabolic process(GO:0006063) glucuronate metabolic process(GO:0019585) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.0 | 0.1 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.0 | 0.0 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.1 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0042196 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.0 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.1 | GO:0061518 | microglial cell activation involved in immune response(GO:0002282) macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:0060849 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.1 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.1 | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.1 | GO:0090155 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.1 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.0 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.0 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) peptidyl-glycine modification(GO:0018201) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.0 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.0 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.0 | 0.0 | GO:0090172 | microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.0 | 0.0 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.0 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.0 | GO:2000318 | positive regulation of memory T cell differentiation(GO:0043382) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:1903925 | response to bisphenol A(GO:1903925) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) |
| 0.0 | 0.1 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.0 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.0 | 0.0 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.0 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.0 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) negative regulation of connective tissue replacement(GO:1905204) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 1.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.0 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.0 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.2 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.0 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.0 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.0 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.0 | GO:0033575 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 | 0.2 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.0 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.0 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.0 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) |
| 0.0 | 0.0 | GO:0035600 | tRNA methylthiolation(GO:0035600) |
| 0.0 | 0.1 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.0 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.0 | GO:2000551 | negative regulation of chemokine biosynthetic process(GO:0045079) regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.0 | 0.1 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.0 | GO:1904976 | cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.0 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.0 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:1903349 | omegasome membrane(GO:1903349) omegasome(GO:1990462) |
| 0.0 | 0.1 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.0 | 0.3 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:0032173 | septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.0 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.0 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.0 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.0 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.0 | 0.0 | GO:0045242 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.0 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.0 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.0 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004058 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.1 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.2 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.0 | 0.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 0.0 | 0.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.0 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.0 | GO:0047619 | acylcarnitine hydrolase activity(GO:0047619) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.0 | GO:0008254 | 3'-nucleotidase activity(GO:0008254) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.0 | GO:0008458 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.0 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.3 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.0 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.0 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.0 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.0 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.0 | GO:0035596 | methylthiotransferase activity(GO:0035596) transferase activity, transferring alkylthio groups(GO:0050497) |
| 0.0 | 0.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.0 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.0 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |