Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
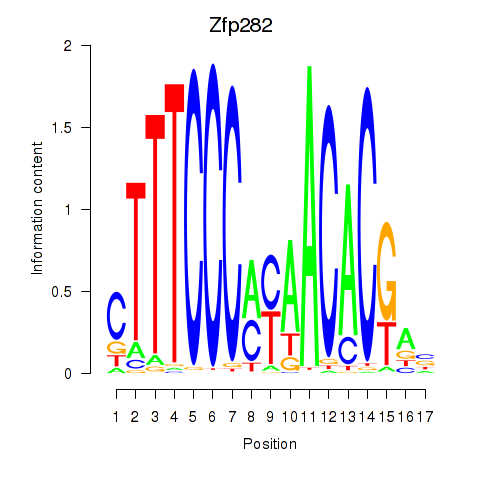
Results for Zfp282
Z-value: 0.21
Transcription factors associated with Zfp282
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp282
|
ENSRNOG00000026981 | zinc finger protein 282 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp282 | rn6_v1_chr4_+_77554269_77554269 | -0.77 | 1.3e-01 | Click! |
Activity profile of Zfp282 motif
Sorted Z-values of Zfp282 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_15301998 | 0.09 |
ENSRNOT00000016400
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr10_-_90312386 | 0.07 |
ENSRNOT00000028445
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr2_+_187447501 | 0.06 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr10_+_16542882 | 0.04 |
ENSRNOT00000028146
|
Stc2
|
stanniocalcin 2 |
| chr2_+_190073815 | 0.04 |
ENSRNOT00000015473
|
S100a8
|
S100 calcium binding protein A8 |
| chr4_-_82399458 | 0.04 |
ENSRNOT00000072532
|
AABR07060588.2
|
|
| chr16_-_48921242 | 0.04 |
ENSRNOT00000041596
|
Cenpu
|
centromere protein U |
| chr13_-_88497901 | 0.03 |
ENSRNOT00000058560
|
Uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr1_-_154111725 | 0.03 |
ENSRNOT00000055488
|
Ccdc81
|
coiled-coil domain containing 81 |
| chr4_+_30313102 | 0.03 |
ENSRNOT00000012657
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr10_+_14828597 | 0.03 |
ENSRNOT00000025434
|
Tekt4
|
tektin 4 |
| chr14_-_44375804 | 0.03 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr9_+_94721681 | 0.03 |
ENSRNOT00000022818
|
Neu2
|
neuraminidase 2 |
| chr7_-_61798729 | 0.03 |
ENSRNOT00000010283
|
Dyrk2
|
dual specificity tyrosine phosphorylation regulated kinase 2 |
| chr1_+_85213652 | 0.03 |
ENSRNOT00000092044
|
Nccrp1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr4_+_157416904 | 0.03 |
ENSRNOT00000059520
|
AC115420.1
|
|
| chr19_+_25946979 | 0.03 |
ENSRNOT00000004027
|
Gadd45gip1
|
GADD45G interacting protein 1 |
| chr15_+_12570062 | 0.02 |
ENSRNOT00000010675
|
Thoc7
|
THO complex 7 |
| chr15_+_104186792 | 0.02 |
ENSRNOT00000014182
|
Dnajc3
|
DnaJ heat shock protein family (Hsp40) member C3 |
| chr8_+_118926478 | 0.02 |
ENSRNOT00000028426
|
Ccdc12
|
coiled-coil domain containing 12 |
| chr4_+_118167294 | 0.02 |
ENSRNOT00000022367
|
LOC687679
|
similar to small nuclear ribonucleoprotein polypeptide G |
| chr5_+_79367663 | 0.02 |
ENSRNOT00000011508
|
Atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr1_+_86948918 | 0.02 |
ENSRNOT00000084839
|
Sirt2
|
sirtuin 2 |
| chr8_+_117694605 | 0.02 |
ENSRNOT00000027994
|
Col7a1
|
collagen type VII alpha 1 chain |
| chr14_+_113968563 | 0.02 |
ENSRNOT00000006085
|
Clhc1
|
clathrin heavy chain linker domain containing 1 |
| chr8_-_68312909 | 0.02 |
ENSRNOT00000066106
|
RGD1309779
|
similar to ENSANGP00000021391 |
| chr1_+_247110216 | 0.02 |
ENSRNOT00000016000
|
Cdc37l1
|
cell division cycle 37-like 1 |
| chr1_-_86948845 | 0.02 |
ENSRNOT00000027212
|
Nfkbib
|
NFKB inhibitor beta |
| chr16_+_5082239 | 0.02 |
ENSRNOT00000074596
|
LOC102547963
|
leucine-rich repeat and transmembrane domain-containing protein 1-like |
| chr7_-_70480724 | 0.02 |
ENSRNOT00000090057
|
Dtx3
|
deltex E3 ubiquitin ligase 3 |
| chr16_+_4997461 | 0.02 |
ENSRNOT00000071042
|
LOC102547963
|
leucine-rich repeat and transmembrane domain-containing protein 1-like |
| chr14_+_44524753 | 0.01 |
ENSRNOT00000076245
|
Rpl9
|
ribosomal protein L9 |
| chr14_+_44524416 | 0.01 |
ENSRNOT00000038927
|
Rpl9
|
ribosomal protein L9 |
| chr2_+_240527130 | 0.01 |
ENSRNOT00000031348
|
Slc9b1
|
solute carrier family 9 member B1 |
| chr13_-_89545182 | 0.01 |
ENSRNOT00000078402
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr1_-_102970950 | 0.01 |
ENSRNOT00000018194
|
Tsg101
|
tumor susceptibility 101 |
| chr3_-_151224123 | 0.01 |
ENSRNOT00000026091
|
Trpc4ap
|
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr15_+_57290849 | 0.01 |
ENSRNOT00000014909
|
Cpb2
|
carboxypeptidase B2 |
| chr20_-_3397039 | 0.01 |
ENSRNOT00000001084
ENSRNOT00000085259 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr15_-_34693034 | 0.01 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr8_-_90664554 | 0.01 |
ENSRNOT00000039917
ENSRNOT00000074896 ENSRNOT00000072225 |
Hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr17_-_34945317 | 0.01 |
ENSRNOT00000090457
|
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr2_-_165641573 | 0.01 |
ENSRNOT00000013987
|
Trim59
|
tripartite motif-containing 59 |
| chr14_-_44524252 | 0.01 |
ENSRNOT00000003779
|
Lias
|
lipoic acid synthetase |
| chr8_-_53816447 | 0.01 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr6_-_21880003 | 0.01 |
ENSRNOT00000006505
|
Ttc27
|
tetratricopeptide repeat domain 27 |
| chr18_+_40962146 | 0.01 |
ENSRNOT00000035656
|
Arl14epl
|
ADP ribosylation factor like GTPase 14 effector protein like |
| chrX_+_78042859 | 0.01 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr14_-_84662143 | 0.01 |
ENSRNOT00000057529
ENSRNOT00000080078 |
Hormad2
|
HORMA domain containing 2 |
| chr3_+_122836446 | 0.01 |
ENSRNOT00000028816
|
Ebf4
|
early B-cell factor 4 |
| chr5_+_28395296 | 0.01 |
ENSRNOT00000009375
|
Tmem55a
|
transmembrane protein 55A |
| chr9_+_4269160 | 0.01 |
ENSRNOT00000050420
|
Sult1c2
|
sulfotransferase family 1C member 2 |
| chr8_+_50563047 | 0.00 |
ENSRNOT00000025149
|
Zfp259
|
zinc finger protein 259 |
| chr3_+_139894331 | 0.00 |
ENSRNOT00000064695
|
Rin2
|
Ras and Rab interactor 2 |
| chr1_+_257157264 | 0.00 |
ENSRNOT00000067149
ENSRNOT00000079219 |
Plce1
|
phospholipase C, epsilon 1 |
| chr16_-_61091169 | 0.00 |
ENSRNOT00000016328
|
Dusp4
|
dual specificity phosphatase 4 |
| chr20_+_29558689 | 0.00 |
ENSRNOT00000085229
|
Ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr3_+_121235119 | 0.00 |
ENSRNOT00000023419
|
Mertk
|
MER proto-oncogene, tyrosine kinase |
| chr17_-_5463158 | 0.00 |
ENSRNOT00000090032
|
Naa35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr1_+_247228061 | 0.00 |
ENSRNOT00000020809
|
Rcl1
|
RNA terminal phosphate cyclase-like 1 |
| chrX_+_37329779 | 0.00 |
ENSRNOT00000038352
ENSRNOT00000088802 |
Pdha1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chrX_-_88321834 | 0.00 |
ENSRNOT00000020364
|
LOC100909548
|
cation-dependent mannose-6-phosphate receptor-like |
| chrX_-_114658931 | 0.00 |
ENSRNOT00000006008
|
Chrdl1
|
chordin-like 1 |
| chr7_-_116063078 | 0.00 |
ENSRNOT00000076932
ENSRNOT00000035496 |
Gml
|
glycosylphosphatidylinositol anchored molecule like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp282
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0010037 | response to carbon dioxide(GO:0010037) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |