Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
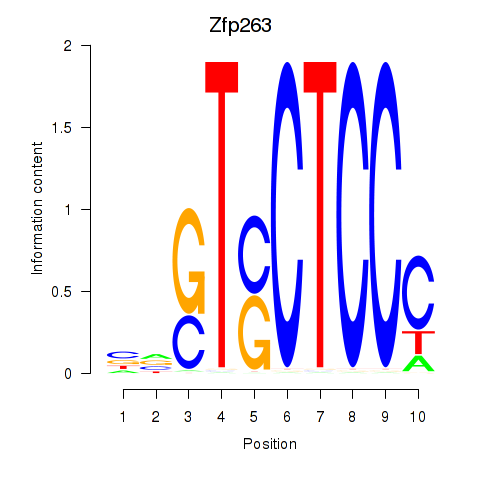
Results for Zfp263
Z-value: 0.71
Transcription factors associated with Zfp263
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp263
|
ENSRNOG00000007678 | zinc finger protein 263 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp263 | rn6_v1_chr10_-_12030416_12030416 | 0.77 | 1.2e-01 | Click! |
Activity profile of Zfp263 motif
Sorted Z-values of Zfp263 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_123445613 | 0.40 |
ENSRNOT00000070937
|
Shisa8
|
shisa family member 8 |
| chr18_+_27657628 | 0.38 |
ENSRNOT00000026303
|
Egr1
|
early growth response 1 |
| chr7_-_119896925 | 0.33 |
ENSRNOT00000010609
|
Elfn2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
| chr10_+_85301875 | 0.30 |
ENSRNOT00000080935
|
Socs7
|
suppressor of cytokine signaling 7 |
| chr17_+_4846789 | 0.29 |
ENSRNOT00000073271
|
Gas1
|
growth arrest-specific 1 |
| chr14_+_19866408 | 0.29 |
ENSRNOT00000060465
|
Adamts3
|
ADAM metallopeptidase with thrombospondin type 1, motif 3 |
| chr15_+_51756978 | 0.28 |
ENSRNOT00000024067
|
Egr3
|
early growth response 3 |
| chr3_+_155160481 | 0.28 |
ENSRNOT00000021133
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr16_-_81434038 | 0.27 |
ENSRNOT00000067508
|
Rasa3
|
RAS p21 protein activator 3 |
| chr7_-_117151256 | 0.25 |
ENSRNOT00000078846
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr7_-_11648322 | 0.24 |
ENSRNOT00000026871
|
Gadd45b
|
growth arrest and DNA-damage-inducible, beta |
| chr7_-_142210738 | 0.24 |
ENSRNOT00000006095
|
Pou6f1
|
POU class 6 homeobox 1 |
| chr19_+_10119253 | 0.22 |
ENSRNOT00000017971
|
Zfp319
|
zinc finger protein 319 |
| chr12_-_2555164 | 0.21 |
ENSRNOT00000084460
ENSRNOT00000061821 |
Map2k7
|
mitogen activated protein kinase kinase 7 |
| chr19_-_26094756 | 0.20 |
ENSRNOT00000067780
|
Junb
|
JunB proto-oncogene, AP-1 transcription factor subunit |
| chr3_-_58181971 | 0.20 |
ENSRNOT00000002076
|
Dlx2
|
distal-less homeobox 2 |
| chr10_+_67325347 | 0.19 |
ENSRNOT00000079002
ENSRNOT00000085914 |
Suz12
|
suppressor of zeste 12 homolog (Drosophila) |
| chr6_+_132702448 | 0.19 |
ENSRNOT00000005743
|
Yy1
|
YY1 transcription factor |
| chr13_+_97838361 | 0.18 |
ENSRNOT00000003641
|
Cnst
|
consortin, connexin sorting protein |
| chr1_+_81208212 | 0.18 |
ENSRNOT00000026288
|
Lypd5
|
Ly6/Plaur domain containing 5 |
| chr15_-_48445588 | 0.18 |
ENSRNOT00000077197
ENSRNOT00000018583 |
Extl3
|
exostosin-like glycosyltransferase 3 |
| chr1_+_165724451 | 0.17 |
ENSRNOT00000025827
|
Fam168a
|
family with sequence similarity 168, member A |
| chr13_+_98615287 | 0.17 |
ENSRNOT00000004032
|
Itpkb
|
inositol-trisphosphate 3-kinase B |
| chr10_-_74679858 | 0.17 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr10_+_61685645 | 0.17 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr7_-_47586137 | 0.17 |
ENSRNOT00000006086
|
Tmtc2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr1_-_220265772 | 0.16 |
ENSRNOT00000027119
|
Npas4
|
neuronal PAS domain protein 4 |
| chr18_+_30885789 | 0.15 |
ENSRNOT00000044434
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
| chr13_+_110398800 | 0.15 |
ENSRNOT00000064492
|
Lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr10_-_110101872 | 0.15 |
ENSRNOT00000071893
|
Ccdc57
|
coiled-coil domain containing 57 |
| chr12_-_21761487 | 0.15 |
ENSRNOT00000082910
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr6_-_110904288 | 0.15 |
ENSRNOT00000014645
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr10_-_13580821 | 0.15 |
ENSRNOT00000009735
|
Ntn3
|
netrin 3 |
| chr2_-_189096785 | 0.15 |
ENSRNOT00000028200
|
Chrnb2
|
cholinergic receptor nicotinic beta 2 subunit |
| chr14_+_16491573 | 0.14 |
ENSRNOT00000002995
|
Sowahb
|
sosondowah ankyrin repeat domain family member B |
| chr18_-_38088457 | 0.14 |
ENSRNOT00000077814
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr8_+_111600532 | 0.14 |
ENSRNOT00000081952
|
Rab6b
|
RAB6B, member RAS oncogene family |
| chr1_+_190666149 | 0.14 |
ENSRNOT00000089361
|
LOC102547219
|
uncharacterized LOC102547219 |
| chr12_+_29266602 | 0.14 |
ENSRNOT00000076194
|
Wbscr17
|
Williams-Beuren syndrome chromosome region 17 |
| chr10_+_93520132 | 0.14 |
ENSRNOT00000055137
|
Mrc2
|
mannose receptor, C type 2 |
| chr15_-_43542939 | 0.14 |
ENSRNOT00000012996
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr15_-_2966576 | 0.14 |
ENSRNOT00000070893
ENSRNOT00000017383 |
Kat6b
|
lysine acetyltransferase 6B |
| chr17_-_18592750 | 0.14 |
ENSRNOT00000065742
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr18_-_37776453 | 0.13 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chrX_-_74706214 | 0.13 |
ENSRNOT00000040637
|
Slc16a2
|
solute carrier family 16 member 2 |
| chr7_-_139907640 | 0.13 |
ENSRNOT00000045473
|
Zfp641
|
zinc finger protein 641 |
| chr6_+_54488038 | 0.13 |
ENSRNOT00000084661
ENSRNOT00000005637 |
Snx13
|
sorting nexin 13 |
| chr13_-_98772968 | 0.13 |
ENSRNOT00000057735
|
Stum
|
stum, mechanosensory transduction mediator homolog |
| chr1_+_185356975 | 0.13 |
ENSRNOT00000086681
|
Plekha7
|
pleckstrin homology domain containing A7 |
| chr9_+_16427589 | 0.13 |
ENSRNOT00000091809
|
Gltscr1l
|
GLTSCR1-like |
| chr2_+_174542667 | 0.13 |
ENSRNOT00000076793
|
Fstl5
|
follistatin-like 5 |
| chr9_-_82699551 | 0.13 |
ENSRNOT00000020673
|
Obsl1
|
obscurin-like 1 |
| chr5_+_58855773 | 0.13 |
ENSRNOT00000072869
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr12_+_22434814 | 0.13 |
ENSRNOT00000076829
|
Slc12a9
|
solute carrier family 12, member 9 |
| chr10_-_103848035 | 0.13 |
ENSRNOT00000029001
|
Fads6
|
fatty acid desaturase 6 |
| chr18_+_30869628 | 0.13 |
ENSRNOT00000060470
|
Pcdhgb4
|
protocadherin gamma subfamily B, 4 |
| chr7_+_41475163 | 0.12 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr1_-_57327379 | 0.12 |
ENSRNOT00000080429
|
Dll1
|
delta like canonical Notch ligand 1 |
| chr8_+_78858570 | 0.12 |
ENSRNOT00000089335
|
Zfp280d
|
zinc finger protein 280D |
| chr3_-_107760550 | 0.12 |
ENSRNOT00000077091
ENSRNOT00000051638 |
Meis2
|
Meis homeobox 2 |
| chr1_-_81450094 | 0.12 |
ENSRNOT00000035356
|
Zfp575
|
zinc finger protein 575 |
| chr10_+_103703404 | 0.12 |
ENSRNOT00000086469
|
Rab37
|
RAB37, member RAS oncogene family |
| chr14_-_72380330 | 0.12 |
ENSRNOT00000082653
ENSRNOT00000081878 ENSRNOT00000006727 |
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr1_+_247688789 | 0.11 |
ENSRNOT00000021783
|
Ric1
|
RIC1 homolog, RAB6A GEF complex partner 1 |
| chr15_+_15275541 | 0.11 |
ENSRNOT00000012153
|
Cadps
|
calcium dependent secretion activator |
| chr7_+_142575672 | 0.11 |
ENSRNOT00000080923
ENSRNOT00000008160 |
Scn8a
|
sodium voltage-gated channel alpha subunit 8 |
| chr15_+_34167945 | 0.11 |
ENSRNOT00000032252
|
Carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr10_+_54246250 | 0.11 |
ENSRNOT00000004880
|
Rcvrn
|
recoverin |
| chr4_-_183293999 | 0.11 |
ENSRNOT00000079695
|
Ipo8
|
importin 8 |
| chr3_+_72540538 | 0.11 |
ENSRNOT00000012379
|
Aplnr
|
apelin receptor |
| chr7_+_143707237 | 0.11 |
ENSRNOT00000074212
|
Tns2
|
tensin 2 |
| chr1_+_46835971 | 0.11 |
ENSRNOT00000066798
|
Synj2
|
synaptojanin 2 |
| chr2_+_201289357 | 0.11 |
ENSRNOT00000067358
|
Tbx15
|
T-box 15 |
| chr12_-_6956914 | 0.10 |
ENSRNOT00000072129
ENSRNOT00000001210 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr11_-_90406797 | 0.10 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
| chrX_+_105239840 | 0.10 |
ENSRNOT00000039864
|
Drp2
|
dystrophin related protein 2 |
| chrX_-_15693473 | 0.10 |
ENSRNOT00000013758
|
Prickle3
|
prickle planar cell polarity protein 3 |
| chr1_-_131454689 | 0.10 |
ENSRNOT00000014152
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr14_+_75852060 | 0.10 |
ENSRNOT00000075975
|
Hs3st1
|
heparan sulfate-glucosamine 3-sulfotransferase 1 |
| chr17_+_27496353 | 0.10 |
ENSRNOT00000077994
ENSRNOT00000019040 ENSRNOT00000089736 |
Ssr1
|
signal sequence receptor subunit 1 |
| chr15_+_23665202 | 0.10 |
ENSRNOT00000089226
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr7_-_12777901 | 0.10 |
ENSRNOT00000017245
|
Tmem259
|
transmembrane protein 259 |
| chr13_-_86671515 | 0.10 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr5_-_59553416 | 0.10 |
ENSRNOT00000090490
|
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr5_-_137104993 | 0.10 |
ENSRNOT00000027271
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F |
| chr4_+_120671489 | 0.10 |
ENSRNOT00000029125
|
Mgll
|
monoglyceride lipase |
| chr9_-_43022998 | 0.10 |
ENSRNOT00000063781
ENSRNOT00000089843 |
Lman2l
|
lectin, mannose-binding 2-like |
| chr12_+_21721837 | 0.10 |
ENSRNOT00000073353
|
Mepce
|
methylphosphate capping enzyme |
| chr2_+_95320283 | 0.10 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr7_-_140546908 | 0.09 |
ENSRNOT00000077502
|
Kmt2d
|
lysine methyltransferase 2D |
| chrX_+_71155601 | 0.09 |
ENSRNOT00000076453
ENSRNOT00000048521 |
Foxo4
|
forkhead box O4 |
| chrX_-_157095274 | 0.09 |
ENSRNOT00000084088
|
Abcd1
|
ATP binding cassette subfamily D member 1 |
| chr11_+_83868655 | 0.09 |
ENSRNOT00000072402
|
NEWGENE_621438
|
thrombopoietin |
| chr15_+_108908607 | 0.09 |
ENSRNOT00000089455
|
Zic2
|
Zic family member 2 |
| chr3_+_161272385 | 0.09 |
ENSRNOT00000021052
|
Zswim3
|
zinc finger, SWIM-type containing 3 |
| chr5_+_1417478 | 0.09 |
ENSRNOT00000008153
ENSRNOT00000085564 |
Jph1
|
junctophilin 1 |
| chr10_-_72142533 | 0.09 |
ENSRNOT00000030885
|
Mrm1
|
mitochondrial rRNA methyltransferase 1 |
| chr20_-_29199224 | 0.09 |
ENSRNOT00000071477
|
Mcu
|
mitochondrial calcium uniporter |
| chr4_-_66002444 | 0.08 |
ENSRNOT00000018645
|
Zc3hav1l
|
zinc finger CCCH-type containing, antiviral 1 like |
| chr1_-_64147251 | 0.08 |
ENSRNOT00000088502
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr2_+_198721724 | 0.08 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chrX_+_50599060 | 0.08 |
ENSRNOT00000058420
|
AABR07038477.1
|
|
| chr7_-_28040510 | 0.08 |
ENSRNOT00000005674
|
Ascl1
|
achaete-scute family bHLH transcription factor 1 |
| chr15_-_4022223 | 0.08 |
ENSRNOT00000081997
|
Zswim8
|
zinc finger, SWIM-type containing 8 |
| chr8_+_49737798 | 0.08 |
ENSRNOT00000022476
|
Dscaml1
|
DS cell adhesion molecule-like 1 |
| chr19_-_57192095 | 0.08 |
ENSRNOT00000058080
|
Pgbd5
|
piggyBac transposable element derived 5 |
| chr18_+_76559811 | 0.07 |
ENSRNOT00000084621
|
Pard6g
|
par-6 family cell polarity regulator gamma |
| chr8_-_61290240 | 0.07 |
ENSRNOT00000023084
|
Lingo1
|
leucine rich repeat and Ig domain containing 1 |
| chr2_+_230901126 | 0.07 |
ENSRNOT00000016026
ENSRNOT00000015564 ENSRNOT00000068198 |
Camk2d
|
calcium/calmodulin-dependent protein kinase II delta |
| chr1_-_226152524 | 0.07 |
ENSRNOT00000027756
|
Fads2
|
fatty acid desaturase 2 |
| chr5_-_75319189 | 0.07 |
ENSRNOT00000047200
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr1_+_81328183 | 0.07 |
ENSRNOT00000074985
ENSRNOT00000075167 |
Plaur
|
plasminogen activator, urokinase receptor |
| chr1_+_221644867 | 0.07 |
ENSRNOT00000054830
|
Ehd1
|
EH-domain containing 1 |
| chr19_+_39543242 | 0.07 |
ENSRNOT00000018837
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr5_+_169452493 | 0.07 |
ENSRNOT00000014444
|
Gpr153
|
G protein-coupled receptor 153 |
| chr4_+_155653718 | 0.07 |
ENSRNOT00000065419
|
Foxj2
|
forkhead box J2 |
| chr3_+_148510779 | 0.07 |
ENSRNOT00000012156
|
Xkr7
|
XK related 7 |
| chr1_-_100810522 | 0.07 |
ENSRNOT00000027260
|
Atf5
|
activating transcription factor 5 |
| chr11_+_30550141 | 0.07 |
ENSRNOT00000002866
|
Hunk
|
hormonally upregulated Neu-associated kinase |
| chr15_-_33589522 | 0.07 |
ENSRNOT00000022514
|
Efs
|
embryonal Fyn-associated substrate |
| chr8_-_47393503 | 0.07 |
ENSRNOT00000059868
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr7_-_123767797 | 0.07 |
ENSRNOT00000012699
|
Tcf20
|
transcription factor 20 |
| chr5_+_151211342 | 0.07 |
ENSRNOT00000067939
|
Ahdc1
|
AT hook, DNA binding motif, containing 1 |
| chr15_-_33537146 | 0.07 |
ENSRNOT00000019992
|
Ppp1r3e
|
protein phosphatase 1, regulatory subunit 3E |
| chr7_-_129724303 | 0.06 |
ENSRNOT00000006034
|
Brd1
|
bromodomain containing 1 |
| chr10_-_82197520 | 0.06 |
ENSRNOT00000092024
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr4_+_157523110 | 0.06 |
ENSRNOT00000081640
|
Zfp384
|
zinc finger protein 384 |
| chr8_+_111495331 | 0.06 |
ENSRNOT00000091275
ENSRNOT00000012162 |
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chr10_+_103934797 | 0.06 |
ENSRNOT00000035865
|
Cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr1_+_251145253 | 0.06 |
ENSRNOT00000083555
ENSRNOT00000014979 |
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr3_+_64190973 | 0.06 |
ENSRNOT00000011342
|
AABR07052587.1
|
|
| chr5_-_75319765 | 0.06 |
ENSRNOT00000085698
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr8_+_39848448 | 0.06 |
ENSRNOT00000012248
|
Hepacam
|
hepatocyte cell adhesion molecule |
| chr2_+_125752130 | 0.06 |
ENSRNOT00000038703
|
Fat4
|
FAT atypical cadherin 4 |
| chr1_-_89042176 | 0.06 |
ENSRNOT00000080842
|
Kmt2b
|
lysine methyltransferase 2B |
| chrX_+_105239620 | 0.06 |
ENSRNOT00000085693
|
Drp2
|
dystrophin related protein 2 |
| chr15_+_57340579 | 0.06 |
ENSRNOT00000015467
|
Zc3h13
|
zinc finger CCCH type containing 13 |
| chr2_+_113984824 | 0.06 |
ENSRNOT00000086399
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr3_-_57119001 | 0.06 |
ENSRNOT00000083171
|
Tlk1
|
tousled-like kinase 1 |
| chr1_-_207171760 | 0.06 |
ENSRNOT00000047069
|
Fam196a
|
family with sequence similarity 196, member A |
| chr1_+_226077120 | 0.06 |
ENSRNOT00000027611
|
Rab3il1
|
RAB3A interacting protein-like 1 |
| chrX_+_140878216 | 0.06 |
ENSRNOT00000075840
ENSRNOT00000077026 |
Zic3
|
Zic family member 3 |
| chr10_-_88645364 | 0.06 |
ENSRNOT00000030344
|
Rab5c
|
RAB5C, member RAS oncogene family |
| chr1_+_177764445 | 0.06 |
ENSRNOT00000037185
|
Rassf10
|
Ras association domain family member 10 |
| chr5_-_159662754 | 0.05 |
ENSRNOT00000045226
|
Szrd1
|
SUZ RNA binding domain containing 1 |
| chr2_+_199199845 | 0.05 |
ENSRNOT00000020932
|
LOC100909441
|
tubulin alpha-1C chain-like |
| chr10_+_46511271 | 0.05 |
ENSRNOT00000080828
|
Rai1
|
retinoic acid induced 1 |
| chr13_+_99005142 | 0.05 |
ENSRNOT00000004293
|
Acbd3
|
acyl-CoA binding domain containing 3 |
| chr8_-_96203677 | 0.05 |
ENSRNOT00000074515
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr19_+_39067363 | 0.05 |
ENSRNOT00000083515
|
Has3
|
hyaluronan synthase 3 |
| chr10_-_82197848 | 0.05 |
ENSRNOT00000081307
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr8_+_36625733 | 0.05 |
ENSRNOT00000016248
|
Cdon
|
cell adhesion associated, oncogene regulated |
| chr9_-_81565416 | 0.05 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chr10_+_105156822 | 0.05 |
ENSRNOT00000012523
|
Galr2
|
galanin receptor 2 |
| chr1_+_167169442 | 0.05 |
ENSRNOT00000048172
|
LOC102549471
|
short transient receptor potential channel 2-like |
| chr10_+_105499569 | 0.05 |
ENSRNOT00000088457
|
Sphk1
|
sphingosine kinase 1 |
| chr11_-_25078740 | 0.05 |
ENSRNOT00000002109
|
Cyyr1
|
cysteine and tyrosine rich 1 |
| chr5_-_173611202 | 0.05 |
ENSRNOT00000047854
|
Agrn
|
agrin |
| chr5_-_147148291 | 0.05 |
ENSRNOT00000085151
|
Azin2
|
antizyme inhibitor 2 |
| chr1_+_221735517 | 0.05 |
ENSRNOT00000028628
ENSRNOT00000044866 |
Sf1
|
splicing factor 1 |
| chr12_-_17972737 | 0.05 |
ENSRNOT00000001783
|
Fam20c
|
FAM20C, golgi associated secretory pathway kinase |
| chr9_+_54212767 | 0.05 |
ENSRNOT00000078073
ENSRNOT00000090026 |
Gls
|
glutaminase |
| chr1_+_226091774 | 0.04 |
ENSRNOT00000027693
|
Fads3
|
fatty acid desaturase 3 |
| chr12_-_24556976 | 0.04 |
ENSRNOT00000041798
|
Bcl7b
|
BCL tumor suppressor 7B |
| chr5_+_144779681 | 0.04 |
ENSRNOT00000016493
|
LOC100294508
|
dyslexia susceptibility 2-like |
| chrX_-_113584459 | 0.04 |
ENSRNOT00000025923
|
Kcne5
|
potassium voltage-gated channel subfamily E regulatory subunit 5 |
| chr10_+_72197977 | 0.04 |
ENSRNOT00000003886
|
Myo19
|
myosin XIX |
| chr15_+_106257650 | 0.04 |
ENSRNOT00000014859
ENSRNOT00000072538 |
Ipo5
|
importin 5 |
| chr2_+_199283909 | 0.04 |
ENSRNOT00000043194
|
Acp6
|
acid phosphatase 6, lysophosphatidic |
| chr8_-_111850075 | 0.04 |
ENSRNOT00000082097
|
Cdv3
|
CDV3 homolog |
| chr16_-_48692476 | 0.04 |
ENSRNOT00000013118
|
Irf2
|
interferon regulatory factor 2 |
| chr20_-_2211995 | 0.04 |
ENSRNOT00000077201
|
Trim26
|
tripartite motif-containing 26 |
| chr13_-_49313940 | 0.04 |
ENSRNOT00000012190
|
Cntn2
|
contactin 2 |
| chr4_-_77706994 | 0.04 |
ENSRNOT00000038517
|
Zfp777
|
zinc finger protein 777 |
| chrX_-_34794589 | 0.04 |
ENSRNOT00000008703
|
Rai2
|
retinoic acid induced 2 |
| chr6_-_92527711 | 0.04 |
ENSRNOT00000007529
|
Nin
|
ninein |
| chr2_-_30577218 | 0.04 |
ENSRNOT00000024674
|
Ocln
|
occludin |
| chr16_+_3051449 | 0.04 |
ENSRNOT00000074069
|
Fam208a
|
family with sequence similarity 208, member A |
| chr10_+_109893700 | 0.04 |
ENSRNOT00000082551
|
Lrrc45
|
leucine rich repeat containing 45 |
| chr8_-_61079526 | 0.04 |
ENSRNOT00000068658
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr14_+_100217289 | 0.03 |
ENSRNOT00000077171
ENSRNOT00000007104 |
Cnrip1
|
cannabinoid receptor interacting protein 1 |
| chr9_-_110936631 | 0.03 |
ENSRNOT00000064431
|
Fbxl17
|
F-box and leucine-rich repeat protein 17 |
| chrX_-_68562873 | 0.03 |
ENSRNOT00000076193
|
Ophn1
|
oligophrenin 1 |
| chr1_-_82409639 | 0.03 |
ENSRNOT00000031326
|
Erich4
|
glutamate-rich 4 |
| chr4_-_56786754 | 0.03 |
ENSRNOT00000050795
|
Kcp
|
kielin/chordin-like protein |
| chr2_-_61949926 | 0.03 |
ENSRNOT00000025966
|
Npr3
|
natriuretic peptide receptor 3 |
| chr11_-_65845418 | 0.03 |
ENSRNOT00000091057
|
Fstl1
|
follistatin-like 1 |
| chr2_+_2456735 | 0.03 |
ENSRNOT00000032974
|
Ell2
|
elongation factor for RNA polymerase II 2 |
| chr6_-_102196138 | 0.03 |
ENSRNOT00000014132
|
Tmem229b
|
transmembrane protein 229B |
| chr5_-_144221263 | 0.03 |
ENSRNOT00000013570
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr14_+_3506339 | 0.03 |
ENSRNOT00000002867
|
Tgfbr3
|
transforming growth factor beta receptor 3 |
| chr7_-_130827152 | 0.03 |
ENSRNOT00000019406
|
Syt10
|
synaptotagmin 10 |
| chr2_-_198852161 | 0.03 |
ENSRNOT00000028815
|
Polr3c
|
RNA polymerase III subunit C |
| chr1_-_198320075 | 0.03 |
ENSRNOT00000081291
|
Taok2
|
TAO kinase 2 |
| chr4_+_22859622 | 0.03 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chr17_+_36334147 | 0.03 |
ENSRNOT00000050261
|
E2f3
|
E2F transcription factor 3 |
| chr11_-_77593171 | 0.03 |
ENSRNOT00000002645
ENSRNOT00000043498 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr6_-_146195819 | 0.03 |
ENSRNOT00000007625
|
Sp4
|
Sp4 transcription factor |
| chr10_+_84955864 | 0.03 |
ENSRNOT00000038572
|
Sp6
|
Sp6 transcription factor |
| chr7_-_62972084 | 0.03 |
ENSRNOT00000064251
|
Msrb3
|
methionine sulfoxide reductase B3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp263
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0098758 | interleukin-1 biosynthetic process(GO:0042222) response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.3 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.2 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.1 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.2 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 0.0 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) cerebellar cortex structural organization(GO:0021698) negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.1 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0060849 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 0.0 | 0.2 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.3 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.1 | GO:0060166 | neuroblast differentiation(GO:0014016) spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) medulla oblongata development(GO:0021550) musculoskeletal movement, spinal reflex action(GO:0050883) olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.2 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.3 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:0010045 | response to nickel cation(GO:0010045) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.1 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.0 | GO:2000541 | positive regulation of synaptic growth at neuromuscular junction(GO:0045887) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) regulation of protein geranylgeranylation(GO:2000539) positive regulation of protein geranylgeranylation(GO:2000541) |
| 0.0 | 0.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.0 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.0 | GO:0060935 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:0045226 | hyaluranon cable assembly(GO:0036118) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.0 | GO:0042662 | negative regulation of mesodermal cell fate specification(GO:0042662) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.0 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0009384 | N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 0.0 | 0.0 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.0 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |