Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
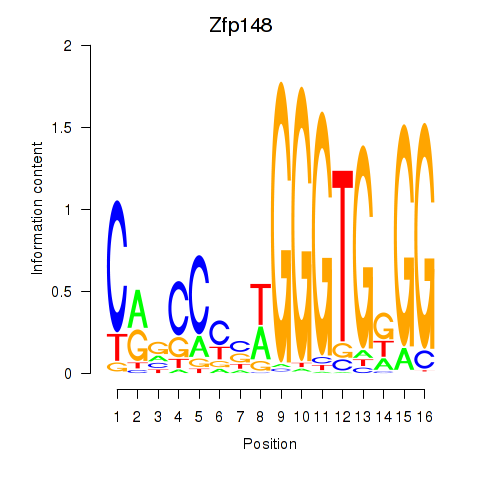
Results for Zfp148
Z-value: 0.43
Transcription factors associated with Zfp148
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp148
|
ENSRNOG00000001789 | zinc finger protein 148 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp148 | rn6_v1_chr11_-_70585053_70585053 | 0.10 | 8.8e-01 | Click! |
Activity profile of Zfp148 motif
Sorted Z-values of Zfp148 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_21353134 | 0.08 |
ENSRNOT00000067898
|
Smim13
|
small integral membrane protein 13 |
| chr5_-_156141537 | 0.08 |
ENSRNOT00000019004
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr1_-_215536770 | 0.07 |
ENSRNOT00000078030
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr13_-_80738634 | 0.07 |
ENSRNOT00000081551
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr5_-_48504511 | 0.07 |
ENSRNOT00000010271
|
Pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr7_-_121232741 | 0.06 |
ENSRNOT00000023196
|
Pdgfb
|
platelet derived growth factor subunit B |
| chr1_+_256101903 | 0.06 |
ENSRNOT00000022384
|
Hhex
|
hematopoietically expressed homeobox |
| chr10_-_90999506 | 0.06 |
ENSRNOT00000034401
|
Gfap
|
glial fibrillary acidic protein |
| chr4_-_100883038 | 0.06 |
ENSRNOT00000041880
|
LOC100364435
|
thymosin, beta 10-like |
| chr11_+_82466071 | 0.06 |
ENSRNOT00000036958
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr1_-_239057732 | 0.06 |
ENSRNOT00000024775
|
Gda
|
guanine deaminase |
| chr12_+_24651314 | 0.06 |
ENSRNOT00000077016
ENSRNOT00000071569 |
Vps37d
|
VPS37D, ESCRT-I subunit |
| chr19_+_14352411 | 0.06 |
ENSRNOT00000067519
|
Hmgxb4
|
HMG-box containing 4 |
| chr8_+_130283542 | 0.06 |
ENSRNOT00000071573
|
Vipr1
|
vasoactive intestinal peptide receptor 1 |
| chr3_-_2689084 | 0.05 |
ENSRNOT00000020926
|
Ptgds
|
prostaglandin D2 synthase |
| chr1_+_82480195 | 0.05 |
ENSRNOT00000028051
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr10_+_103713045 | 0.05 |
ENSRNOT00000004351
|
Slc9a3r1
|
SLC9A3 regulator 1 |
| chr6_-_38228379 | 0.05 |
ENSRNOT00000084924
|
Mycn
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr4_-_100883275 | 0.05 |
ENSRNOT00000022846
|
LOC100364435
|
thymosin, beta 10-like |
| chr4_+_114854458 | 0.05 |
ENSRNOT00000013312
|
LOC500227
|
hypothetical gene supported by BC079424 |
| chr7_-_117680004 | 0.05 |
ENSRNOT00000040422
|
Slc39a4
|
solute carrier family 39 member 4 |
| chr6_+_1657331 | 0.05 |
ENSRNOT00000049672
ENSRNOT00000079864 |
Qpct
|
glutaminyl-peptide cyclotransferase |
| chr1_-_89483988 | 0.05 |
ENSRNOT00000028603
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr3_+_93920447 | 0.05 |
ENSRNOT00000012625
|
Lmo2
|
LIM domain only 2 |
| chr20_+_5040337 | 0.05 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chr10_-_90127600 | 0.05 |
ENSRNOT00000028368
|
Lsm12
|
LSM12 homolog |
| chr11_+_38035450 | 0.05 |
ENSRNOT00000083067
|
Mx2
|
MX dynamin like GTPase 2 |
| chr3_-_112061796 | 0.05 |
ENSRNOT00000056258
|
Pla2g4d
|
phospholipase A2 group IVD |
| chr10_-_88611105 | 0.04 |
ENSRNOT00000024718
|
Dhx58
|
DEXH-box helicase 58 |
| chr2_+_196334626 | 0.04 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr19_+_10119253 | 0.04 |
ENSRNOT00000017971
|
Zfp319
|
zinc finger protein 319 |
| chr7_-_3315856 | 0.04 |
ENSRNOT00000010035
|
Gdf11
|
growth differentiation factor 11 |
| chr2_-_29768750 | 0.04 |
ENSRNOT00000023460
|
Map1b
|
microtubule-associated protein 1B |
| chr12_+_47074200 | 0.04 |
ENSRNOT00000014910
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr10_-_39405311 | 0.04 |
ENSRNOT00000074616
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr11_-_87158597 | 0.04 |
ENSRNOT00000002517
|
Vpreb2
|
pre-B lymphocyte gene 2 |
| chr10_-_72153375 | 0.04 |
ENSRNOT00000030841
|
Dhrs11
|
dehydrogenase/reductase 11 |
| chr1_+_282265370 | 0.04 |
ENSRNOT00000015687
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr16_+_10417185 | 0.04 |
ENSRNOT00000082186
|
Anxa8
|
annexin A8 |
| chr4_-_14490446 | 0.04 |
ENSRNOT00000009132
|
Sema3c
|
semaphorin 3C |
| chr13_-_72744295 | 0.04 |
ENSRNOT00000093366
ENSRNOT00000077157 |
Ier5
|
immediate early response 5 |
| chr19_+_40855433 | 0.04 |
ENSRNOT00000023823
|
Il34
|
interleukin 34 |
| chr1_+_98440186 | 0.04 |
ENSRNOT00000024150
|
Iglon5
|
IgLON family member 5 |
| chr10_+_91710495 | 0.04 |
ENSRNOT00000033276
|
Rprml
|
reprimo-like |
| chr5_+_57472315 | 0.04 |
ENSRNOT00000015575
|
Ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr17_+_15814132 | 0.04 |
ENSRNOT00000032997
|
Susd3
|
sushi domain containing 3 |
| chrX_-_157095274 | 0.04 |
ENSRNOT00000084088
|
Abcd1
|
ATP binding cassette subfamily D member 1 |
| chr1_+_202432366 | 0.04 |
ENSRNOT00000027681
|
Plpp4
|
phospholipid phosphatase 4 |
| chr10_+_59529785 | 0.04 |
ENSRNOT00000064840
ENSRNOT00000065181 |
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr7_+_140716113 | 0.04 |
ENSRNOT00000033450
|
Tuba1c
|
tubulin, alpha 1C |
| chr12_+_52397792 | 0.04 |
ENSRNOT00000085661
ENSRNOT00000056694 ENSRNOT00000056693 ENSRNOT00000056692 ENSRNOT00000081447 |
P2rx2
|
purinergic receptor P2X 2 |
| chr10_-_40805941 | 0.03 |
ENSRNOT00000017588
|
Atox1
|
antioxidant 1 copper chaperone |
| chr3_+_63536166 | 0.03 |
ENSRNOT00000016132
|
Plekha3
|
pleckstrin homology domain containing A3 |
| chr19_+_25969255 | 0.03 |
ENSRNOT00000004370
|
Farsa
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr11_+_38035611 | 0.03 |
ENSRNOT00000087603
ENSRNOT00000002695 |
Mx2
|
MX dynamin like GTPase 2 |
| chr1_+_282134981 | 0.03 |
ENSRNOT00000036203
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr1_+_220869805 | 0.03 |
ENSRNOT00000015962
|
Cfl1
|
cofilin 1 |
| chr11_-_29710849 | 0.03 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr19_-_55510460 | 0.03 |
ENSRNOT00000019820
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr14_+_36687134 | 0.03 |
ENSRNOT00000002879
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr8_-_11887799 | 0.03 |
ENSRNOT00000072420
|
Jrkl
|
JRK-like |
| chr4_-_16130848 | 0.03 |
ENSRNOT00000042914
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr18_+_70733872 | 0.03 |
ENSRNOT00000067018
|
Acaa2
|
acetyl-CoA acyltransferase 2 |
| chr5_-_58163584 | 0.03 |
ENSRNOT00000060594
|
Ccl27
|
C-C motif chemokine ligand 27 |
| chr10_-_85574889 | 0.03 |
ENSRNOT00000072274
|
LOC691153
|
hypothetical protein LOC691153 |
| chr7_+_11054333 | 0.03 |
ENSRNOT00000007202
|
Gna15
|
guanine nucleotide binding protein, alpha 15 |
| chr10_-_13446135 | 0.03 |
ENSRNOT00000084991
|
Kctd5
|
potassium channel tetramerization domain containing 5 |
| chr13_+_89774764 | 0.03 |
ENSRNOT00000005619
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr5_-_139227196 | 0.03 |
ENSRNOT00000050941
|
Foxo6
|
forkhead box O6 |
| chr5_-_154223536 | 0.03 |
ENSRNOT00000012258
|
Pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr3_-_63535991 | 0.03 |
ENSRNOT00000015722
|
Fkbp7
|
FK506 binding protein 7 |
| chr6_+_72461977 | 0.03 |
ENSRNOT00000007887
ENSRNOT00000090716 |
Ap4s1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr2_-_46544457 | 0.03 |
ENSRNOT00000015680
|
Fst
|
follistatin |
| chr20_-_3739968 | 0.03 |
ENSRNOT00000074049
ENSRNOT00000084918 |
Cchcr1
|
coiled-coil alpha-helical rod protein 1 |
| chr10_+_56453877 | 0.03 |
ENSRNOT00000031640
|
Plscr3
|
phospholipid scramblase 3 |
| chr3_-_8852192 | 0.03 |
ENSRNOT00000022786
|
Dolk
|
dolichol kinase |
| chr5_+_137458691 | 0.03 |
ENSRNOT00000075464
ENSRNOT00000076895 |
LOC108348080
|
probable rRNA-processing protein EBP2 |
| chr10_-_6870011 | 0.03 |
ENSRNOT00000003439
|
RGD1309748
|
similar to CG4768-PA |
| chr2_-_144467912 | 0.03 |
ENSRNOT00000040002
|
Ccna1
|
cyclin A1 |
| chr3_-_51297852 | 0.03 |
ENSRNOT00000001607
|
Cobll1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr7_-_109205194 | 0.03 |
ENSRNOT00000093145
|
Zfat
|
zinc finger and AT hook domain containing |
| chr1_-_265506046 | 0.03 |
ENSRNOT00000023963
|
Npm3
|
nucleophosmin/nucleoplasmin, 3 |
| chr7_-_120636563 | 0.03 |
ENSRNOT00000091317
ENSRNOT00000031117 |
Tmem184b
|
transmembrane protein 184B |
| chr9_-_82673898 | 0.02 |
ENSRNOT00000027165
|
Chpf
|
chondroitin polymerizing factor |
| chr9_+_82674202 | 0.02 |
ENSRNOT00000027208
|
Tmem198
|
transmembrane protein 198 |
| chr1_+_157920786 | 0.02 |
ENSRNOT00000014284
|
Fam181b
|
family with sequence similarity 181, member B |
| chr3_-_112789282 | 0.02 |
ENSRNOT00000090680
ENSRNOT00000066181 |
Ttbk2
|
tau tubulin kinase 2 |
| chr2_+_188844073 | 0.02 |
ENSRNOT00000028117
|
Kcnn3
|
potassium calcium-activated channel subfamily N member 3 |
| chr2_-_210413985 | 0.02 |
ENSRNOT00000025036
|
Strip1
|
striatin interacting protein 1 |
| chr2_-_232245319 | 0.02 |
ENSRNOT00000014510
|
LOC691931
|
hypothetical protein LOC691931 |
| chr5_-_57632177 | 0.02 |
ENSRNOT00000080787
ENSRNOT00000092581 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr1_-_215836641 | 0.02 |
ENSRNOT00000080246
|
Igf2
|
insulin-like growth factor 2 |
| chr4_-_114853868 | 0.02 |
ENSRNOT00000090828
|
Wdr54
|
WD repeat domain 54 |
| chr3_-_5402666 | 0.02 |
ENSRNOT00000061276
|
Abo2
|
histo-blood group ABO system transferase 2 |
| chr20_-_3751994 | 0.02 |
ENSRNOT00000072288
|
Pou5f1
|
POU class 5 homeobox 1 |
| chr1_+_101012822 | 0.02 |
ENSRNOT00000027809
|
Rras
|
related RAS viral (r-ras) oncogene homolog |
| chr3_-_177201525 | 0.02 |
ENSRNOT00000022451
|
Rgs19
|
regulator of G-protein signaling 19 |
| chr6_-_55001464 | 0.02 |
ENSRNOT00000006618
|
Ahr
|
aryl hydrocarbon receptor |
| chr8_-_116391158 | 0.02 |
ENSRNOT00000078720
ENSRNOT00000022550 |
Gnai2
|
G protein subunit alpha i2 |
| chr12_-_41448668 | 0.02 |
ENSRNOT00000001856
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr9_+_118842787 | 0.02 |
ENSRNOT00000090512
|
Dlgap1
|
DLG associated protein 1 |
| chr6_+_42568220 | 0.02 |
ENSRNOT00000074829
|
Pdia6
|
protein disulfide isomerase family A, member 6 |
| chr5_-_126666830 | 0.02 |
ENSRNOT00000012102
ENSRNOT00000091165 |
Mrpl37
|
mitochondrial ribosomal protein L37 |
| chr7_-_144880092 | 0.02 |
ENSRNOT00000055281
|
Nfe2
|
nuclear factor, erythroid 2 |
| chr10_+_57057608 | 0.02 |
ENSRNOT00000085810
ENSRNOT00000026335 |
Med11
|
mediator complex subunit 11 |
| chr1_-_215536980 | 0.02 |
ENSRNOT00000027344
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chrX_-_142131545 | 0.02 |
ENSRNOT00000077402
|
Fgf13
|
fibroblast growth factor 13 |
| chr1_+_220668544 | 0.02 |
ENSRNOT00000039842
|
Gal3st3
|
galactose-3-O-sulfotransferase 3 |
| chr17_-_45626212 | 0.02 |
ENSRNOT00000077363
|
Trim27
|
tripartite motif-containing 27 |
| chr7_-_109205354 | 0.02 |
ENSRNOT00000068291
|
Zfat
|
zinc finger and AT hook domain containing |
| chr10_-_19164505 | 0.02 |
ENSRNOT00000009058
|
Foxi1
|
forkhead box I1 |
| chr20_-_3438389 | 0.02 |
ENSRNOT00000001098
ENSRNOT00000090039 |
Flot1
|
flotillin 1 |
| chr17_+_56109549 | 0.02 |
ENSRNOT00000022190
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr2_+_54191538 | 0.02 |
ENSRNOT00000019524
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr5_+_137670295 | 0.02 |
ENSRNOT00000076672
|
Ebna1bp2
|
EBNA1 binding protein 2 |
| chr17_-_9837293 | 0.02 |
ENSRNOT00000091320
ENSRNOT00000022134 |
Prelid1
|
PRELI domain containing 1 |
| chr19_+_2393059 | 0.02 |
ENSRNOT00000018535
|
Cdh11
|
cadherin 11 |
| chr19_+_49695579 | 0.02 |
ENSRNOT00000016963
|
Gan
|
gigaxonin |
| chr3_+_11629556 | 0.02 |
ENSRNOT00000074401
|
St6galnac6
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr8_-_26311350 | 0.02 |
ENSRNOT00000046432
|
Herpud2
|
HERPUD family member 2 |
| chr8_+_22577715 | 0.02 |
ENSRNOT00000048245
ENSRNOT00000041577 |
Carm1
|
coactivator-associated arginine methyltransferase 1 |
| chr17_-_70325855 | 0.02 |
ENSRNOT00000024952
|
Gdi2
|
GDP dissociation inhibitor 2 |
| chr4_-_16130563 | 0.02 |
ENSRNOT00000090240
ENSRNOT00000034969 |
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr10_-_88000423 | 0.02 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr18_-_63488027 | 0.02 |
ENSRNOT00000023763
|
Ptpn2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr8_-_59239954 | 0.01 |
ENSRNOT00000016104
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr19_+_37600148 | 0.01 |
ENSRNOT00000023853
|
Ctcf
|
CCCTC-binding factor |
| chr9_+_81518176 | 0.01 |
ENSRNOT00000078317
ENSRNOT00000019265 ENSRNOT00000088246 ENSRNOT00000084682 |
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr19_-_58399816 | 0.01 |
ENSRNOT00000026843
|
Sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr8_-_125645898 | 0.01 |
ENSRNOT00000036672
|
Rbms3
|
RNA binding motif, single stranded interacting protein 3 |
| chr2_+_187282000 | 0.01 |
ENSRNOT00000065211
|
Hdgf
|
hepatoma-derived growth factor |
| chr7_+_141054494 | 0.01 |
ENSRNOT00000088176
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr11_+_87204175 | 0.01 |
ENSRNOT00000000306
|
Slc25a1
|
solute carrier family 25 member 1 |
| chr8_-_98738446 | 0.01 |
ENSRNOT00000019860
|
Zic1
|
Zic family member 1 |
| chr1_+_256226179 | 0.01 |
ENSRNOT00000054748
|
Exoc6
|
exocyst complex component 6 |
| chr7_-_70481796 | 0.01 |
ENSRNOT00000006815
|
Dtx3
|
deltex E3 ubiquitin ligase 3 |
| chr1_-_219853329 | 0.01 |
ENSRNOT00000026169
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr15_+_8125526 | 0.01 |
ENSRNOT00000011653
|
Ube2e1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr14_-_18839595 | 0.01 |
ENSRNOT00000078746
|
Cxcl3
|
chemokine (C-X-C motif) ligand 3 |
| chr17_+_8878270 | 0.01 |
ENSRNOT00000015307
|
Pitx1
|
paired-like homeodomain 1 |
| chr8_-_112648880 | 0.01 |
ENSRNOT00000015265
|
Ackr4
|
atypical chemokine receptor 4 |
| chr6_+_28235695 | 0.01 |
ENSRNOT00000047210
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr3_+_171743929 | 0.01 |
ENSRNOT00000079113
ENSRNOT00000064400 |
Rab22a
|
RAB22A, member RAS oncogene family |
| chr7_+_71057911 | 0.01 |
ENSRNOT00000037218
|
Rdh16
|
retinol dehydrogenase 16 (all-trans) |
| chr10_-_35800120 | 0.01 |
ENSRNOT00000004361
|
Maml1
|
mastermind-like transcriptional coactivator 1 |
| chr1_-_221269017 | 0.01 |
ENSRNOT00000028363
ENSRNOT00000084396 |
Dpf2
|
double PHD fingers 2 |
| chr16_+_63837216 | 0.01 |
ENSRNOT00000014268
ENSRNOT00000014147 ENSRNOT00000038549 |
Nrg1
|
neuregulin 1 |
| chr7_-_136853957 | 0.01 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr1_-_82108083 | 0.01 |
ENSRNOT00000027677
ENSRNOT00000084530 |
Gsk3a
|
glycogen synthase kinase 3 alpha |
| chr12_-_18540166 | 0.01 |
ENSRNOT00000001792
|
Il3ra
|
interleukin 3 receptor subunit alpha |
| chr10_-_84698886 | 0.01 |
ENSRNOT00000067542
|
Nfe2l1
|
nuclear factor, erythroid 2-like 1 |
| chr15_+_12569235 | 0.01 |
ENSRNOT00000083787
|
Thoc7
|
THO complex 7 |
| chr15_+_33555640 | 0.01 |
ENSRNOT00000021096
|
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr2_-_123281856 | 0.01 |
ENSRNOT00000079745
|
Ccna2
|
cyclin A2 |
| chr2_+_211078334 | 0.01 |
ENSRNOT00000049127
|
Sort1
|
sortilin 1 |
| chr14_-_35652709 | 0.01 |
ENSRNOT00000003080
|
Gsx2
|
GS homeobox 2 |
| chr7_+_143060597 | 0.01 |
ENSRNOT00000087481
|
Krt7
|
keratin 7 |
| chr11_+_86903122 | 0.01 |
ENSRNOT00000048063
|
Zdhhc8
|
zinc finger, DHHC-type containing 8 |
| chr3_-_172537877 | 0.01 |
ENSRNOT00000072069
|
Ctsz
|
cathepsin Z |
| chr14_+_42402081 | 0.01 |
ENSRNOT00000092933
|
Bend4
|
BEN domain containing 4 |
| chr3_+_92969141 | 0.01 |
ENSRNOT00000009797
|
Apip
|
APAF1 interacting protein |
| chr18_+_59748444 | 0.01 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr17_+_82430836 | 0.01 |
ENSRNOT00000083164
|
Malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr4_-_115453659 | 0.01 |
ENSRNOT00000065847
|
Tex261
|
testis expressed 261 |
| chr1_+_199360645 | 0.01 |
ENSRNOT00000026527
|
Kat8
|
lysine acetyltransferase 8 |
| chr8_-_44327551 | 0.01 |
ENSRNOT00000083939
|
Gramd1b
|
GRAM domain containing 1B |
| chr3_-_4394428 | 0.01 |
ENSRNOT00000044775
|
Abo
|
ABO, alpha 1-3-N-acetylgalactosaminyltransferase and alpha 1-3-galactosyltransferase |
| chr14_+_84465515 | 0.01 |
ENSRNOT00000035591
|
Osm
|
oncostatin M |
| chrX_-_15761932 | 0.01 |
ENSRNOT00000015641
ENSRNOT00000091146 |
Foxp3
|
forkhead box P3 |
| chr5_-_58113553 | 0.01 |
ENSRNOT00000046589
|
Arid3c
|
AT-rich interaction domain 3C |
| chr5_-_151977636 | 0.01 |
ENSRNOT00000009173
|
Arid1a
|
AT-rich interaction domain 1A |
| chr20_-_7930929 | 0.01 |
ENSRNOT00000000607
|
Tead3
|
TEA domain transcription factor 3 |
| chr10_+_89376530 | 0.01 |
ENSRNOT00000028089
|
Rnd2
|
Rho family GTPase 2 |
| chr3_+_93968855 | 0.01 |
ENSRNOT00000014478
|
Fbxo3
|
F-box protein 3 |
| chr1_-_2073896 | 0.01 |
ENSRNOT00000061999
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr6_-_86822094 | 0.01 |
ENSRNOT00000006531
|
Fkbp3
|
FK506 binding protein 3 |
| chr14_+_84306466 | 0.01 |
ENSRNOT00000006116
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr4_-_67162808 | 0.01 |
ENSRNOT00000011915
|
Slc37a3
|
solute carrier family 37, member 3 |
| chr6_+_76675418 | 0.01 |
ENSRNOT00000076169
ENSRNOT00000010948 ENSRNOT00000082286 |
Brms1l
Brms1l
|
breast cancer metastasis-suppressor 1-like breast cancer metastasis-suppressor 1-like |
| chr8_+_26413233 | 0.01 |
ENSRNOT00000063967
ENSRNOT00000008838 |
Sept7
|
septin 7 |
| chr8_-_71533069 | 0.01 |
ENSRNOT00000021863
|
Trip4
|
thyroid hormone receptor interactor 4 |
| chr7_-_143738237 | 0.01 |
ENSRNOT00000055320
|
Spryd3
|
SPRY domain containing 3 |
| chr5_+_59063531 | 0.01 |
ENSRNOT00000085311
ENSRNOT00000065909 |
Creb3
|
cAMP responsive element binding protein 3 |
| chr9_-_42805673 | 0.01 |
ENSRNOT00000020558
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr1_-_102849430 | 0.01 |
ENSRNOT00000086856
|
Saa4
|
serum amyloid A4 |
| chr16_-_68968248 | 0.01 |
ENSRNOT00000016885
|
Eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr13_+_85465792 | 0.01 |
ENSRNOT00000082991
ENSRNOT00000005277 |
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr4_-_113988246 | 0.01 |
ENSRNOT00000013128
|
LOC103692166
|
WD repeat-containing protein 54 |
| chr14_+_3058993 | 0.01 |
ENSRNOT00000002807
|
Gfi1
|
growth factor independent 1 transcriptional repressor |
| chr8_+_50310405 | 0.01 |
ENSRNOT00000073507
|
Sik3
|
SIK family kinase 3 |
| chr3_+_113818872 | 0.01 |
ENSRNOT00000044158
|
Casc4
|
cancer susceptibility candidate 4 |
| chr10_+_92289107 | 0.01 |
ENSRNOT00000050070
|
Mapt
|
microtubule-associated protein tau |
| chr3_-_92969050 | 0.01 |
ENSRNOT00000088242
|
Pdhx
|
pyruvate dehydrogenase complex, component X |
| chr7_+_141053876 | 0.01 |
ENSRNOT00000084674
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr1_-_81881549 | 0.01 |
ENSRNOT00000027497
|
Atp1a3
|
ATPase Na+/K+ transporting subunit alpha 3 |
| chr13_+_52588917 | 0.01 |
ENSRNOT00000011999
|
Phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr7_-_140546908 | 0.00 |
ENSRNOT00000077502
|
Kmt2d
|
lysine methyltransferase 2D |
| chr16_+_74292438 | 0.00 |
ENSRNOT00000026197
|
Vdac3
|
voltage-dependent anion channel 3 |
| chr10_+_4953879 | 0.00 |
ENSRNOT00000003455
|
Tnp2
|
transition protein 2 |
| chr8_-_94563760 | 0.00 |
ENSRNOT00000032792
|
Snap91
|
synaptosomal-associated protein 91 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp148
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0072254 | metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) |
| 0.0 | 0.1 | GO:0061017 | gall bladder development(GO:0061010) hepatoblast differentiation(GO:0061017) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.0 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.0 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.0 | GO:2000814 | positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0010625 | positive regulation of Schwann cell proliferation(GO:0010625) |
| 0.0 | 0.0 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.0 | GO:0098749 | cerebellar neuron development(GO:0098749) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.0 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |