Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
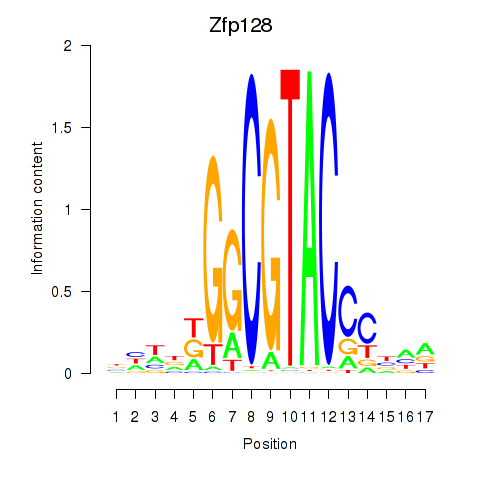
Results for Zfp128
Z-value: 0.22
Transcription factors associated with Zfp128
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp128
|
ENSRNOG00000031113 | zinc finger protein 128 |
|
Zfp128
|
ENSRNOG00000031459 | zinc finger protein 128 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp128 | rn6_v1_chr1_-_65701867_65701867 | 0.24 | 7.0e-01 | Click! |
Activity profile of Zfp128 motif
Sorted Z-values of Zfp128 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_23256158 | 0.09 |
ENSRNOT00000015336
|
Bhmt
|
betaine-homocysteine S-methyltransferase |
| chr10_+_14136883 | 0.07 |
ENSRNOT00000082295
|
AC115181.1
|
|
| chr4_-_150616895 | 0.05 |
ENSRNOT00000073562
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr20_-_31598118 | 0.05 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr5_+_160355833 | 0.04 |
ENSRNOT00000080721
ENSRNOT00000088523 ENSRNOT00000017972 |
Casp9
|
caspase 9 |
| chr1_-_170186462 | 0.04 |
ENSRNOT00000035798
|
Olr202
|
olfactory receptor 202 |
| chr7_-_138039984 | 0.03 |
ENSRNOT00000089806
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr5_+_144581427 | 0.03 |
ENSRNOT00000015227
|
Clspn
|
claspin |
| chr20_-_31597830 | 0.03 |
ENSRNOT00000085877
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr3_-_177089199 | 0.02 |
ENSRNOT00000020873
|
Zfp512b
|
zinc finger protein 512B |
| chr10_+_102136283 | 0.02 |
ENSRNOT00000003735
|
Sstr2
|
somatostatin receptor 2 |
| chr6_+_18877907 | 0.02 |
ENSRNOT00000084222
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr20_-_13581164 | 0.02 |
ENSRNOT00000033298
|
Zfp280b
|
zinc finger protein 280B |
| chr8_-_62215293 | 0.02 |
ENSRNOT00000080448
|
NEWGENE_1306267
|
phosphopantothenoylcysteine decarboxylase |
| chr5_+_155934490 | 0.02 |
ENSRNOT00000077933
|
LOC100911993
|
ubiquitin carboxyl-terminal hydrolase 48-like |
| chr8_-_64154396 | 0.02 |
ENSRNOT00000031262
|
Bbs4
|
Bardet-Biedl syndrome 4 |
| chr5_-_147867583 | 0.01 |
ENSRNOT00000000139
|
Kpna6
|
karyopherin subunit alpha 6 |
| chr2_+_147006830 | 0.01 |
ENSRNOT00000080780
|
AABR07010672.1
|
|
| chr2_-_210839295 | 0.01 |
ENSRNOT00000025994
|
Gstm4
|
glutathione S-transferase mu 4 |
| chr12_-_51341663 | 0.01 |
ENSRNOT00000066892
ENSRNOT00000052202 |
Pitpnb
|
phosphatidylinositol transfer protein, beta |
| chr11_-_45124423 | 0.01 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chr3_-_8690305 | 0.01 |
ENSRNOT00000035002
|
Zer1
|
zyg-11 related, cell cycle regulator |
| chr1_+_162320730 | 0.01 |
ENSRNOT00000035743
|
Kctd21
|
potassium channel tetramerization domain containing 21 |
| chr9_+_98279022 | 0.01 |
ENSRNOT00000038536
|
Rbm44
|
RNA binding motif protein 44 |
| chr7_-_22943159 | 0.01 |
ENSRNOT00000047547
|
Vom2r57
|
vomeronasal 2 receptor, 57 |
| chr18_+_41022552 | 0.01 |
ENSRNOT00000005260
|
Commd10
|
COMM domain containing 10 |
| chr7_-_19088972 | 0.01 |
ENSRNOT00000049266
|
Vom2r57
|
vomeronasal 2 receptor, 57 |
| chr8_-_21384131 | 0.01 |
ENSRNOT00000071010
|
Zfp560
|
zinc finger protein 560 |
| chrX_+_43626480 | 0.01 |
ENSRNOT00000068078
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr11_+_31539016 | 0.01 |
ENSRNOT00000072856
|
Ifnar2
|
interferon alpha and beta receptor subunit 2 |
| chr15_-_34364081 | 0.01 |
ENSRNOT00000027362
|
Tinf2
|
TERF1 interacting nuclear factor 2 |
| chr8_+_13305152 | 0.01 |
ENSRNOT00000012940
|
Mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr6_-_128989812 | 0.01 |
ENSRNOT00000085943
|
LOC100909439
|
ankyrin repeat and SOCS box protein 2-like |
| chr10_+_11240138 | 0.01 |
ENSRNOT00000048687
|
Srl
|
sarcalumenin |
| chr6_-_9459188 | 0.01 |
ENSRNOT00000019894
|
Srbd1
|
S1 RNA binding domain 1 |
| chr14_+_2892753 | 0.01 |
ENSRNOT00000061630
|
Evi5
|
ecotropic viral integration site 5 |
| chr18_+_69549937 | 0.01 |
ENSRNOT00000041227
|
Mex3c
|
mex-3 RNA binding family member C |
| chr3_+_62648447 | 0.01 |
ENSRNOT00000002111
|
Agps
|
alkylglycerone phosphate synthase |
| chr8_-_129371973 | 0.01 |
ENSRNOT00000042614
|
Snrpe
|
small nuclear ribonucleoprotein polypeptide E |
| chr4_+_99398494 | 0.01 |
ENSRNOT00000064374
|
Rnf103
|
ring finger protein 103 |
| chr20_-_43108198 | 0.00 |
ENSRNOT00000000742
|
Hdac2
|
histone deacetylase 2 |
| chr10_+_110631494 | 0.00 |
ENSRNOT00000054915
|
Fn3k
|
fructosamine 3 kinase |
| chr3_-_171893901 | 0.00 |
ENSRNOT00000091966
|
Apcdd1l
|
APC down-regulated 1 like |
| chr12_+_16030788 | 0.00 |
ENSRNOT00000051565
|
Iqce
|
IQ motif containing E |
| chr5_+_151692108 | 0.00 |
ENSRNOT00000086144
|
Fam46b
|
family with sequence similarity 46, member B |
| chr1_-_102845083 | 0.00 |
ENSRNOT00000082289
|
Saa4
|
serum amyloid A4 |
| chr1_-_170471076 | 0.00 |
ENSRNOT00000025159
|
Arfip2
|
ADP-ribosylation factor interacting protein 2 |
| chr8_-_23014499 | 0.00 |
ENSRNOT00000017820
|
Ccdc151
|
coiled-coil domain containing 151 |
| chr8_-_111107599 | 0.00 |
ENSRNOT00000031313
|
Cep63
|
centrosomal protein 63 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp128
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |