Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
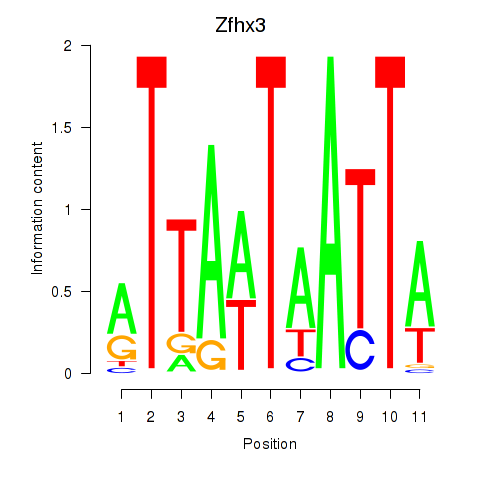
Results for Zfhx3
Z-value: 0.36
Transcription factors associated with Zfhx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfhx3
|
ENSRNOG00000014452 | zinc finger homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfhx3 | rn6_v1_chr19_-_42920344_42920344 | -0.71 | 1.8e-01 | Click! |
Activity profile of Zfhx3 motif
Sorted Z-values of Zfhx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_90818736 | 0.18 |
ENSRNOT00000093480
ENSRNOT00000009083 |
Kdm4c
|
lysine demethylase 4C |
| chr1_-_48891130 | 0.16 |
ENSRNOT00000083884
|
AC135026.1
|
|
| chr2_+_149899836 | 0.15 |
ENSRNOT00000086481
|
RGD1560324
|
similar to hypothetical protein C130079G13 |
| chr8_-_128754514 | 0.14 |
ENSRNOT00000025019
|
Cx3cr1
|
C-X3-C motif chemokine receptor 1 |
| chr6_-_98666007 | 0.10 |
ENSRNOT00000082695
|
AABR07064867.1
|
|
| chr15_-_93765498 | 0.10 |
ENSRNOT00000093297
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr6_-_50941248 | 0.09 |
ENSRNOT00000084533
|
Dus4l
|
dihydrouridine synthase 4-like |
| chrM_+_9870 | 0.09 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr5_+_6373583 | 0.09 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr3_-_55951584 | 0.08 |
ENSRNOT00000036585
|
Fastkd1
|
FAST kinase domains 1 |
| chr9_+_73378057 | 0.07 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr10_-_91661558 | 0.07 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
| chr2_-_177924970 | 0.07 |
ENSRNOT00000029340
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr1_-_166912524 | 0.07 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr7_+_132378273 | 0.06 |
ENSRNOT00000010990
|
LOC690142
|
hypothetical protein LOC690142 |
| chr12_-_29743705 | 0.06 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr14_+_69800156 | 0.05 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr16_-_10802512 | 0.05 |
ENSRNOT00000079554
|
Bmpr1a
|
bone morphogenetic protein receptor type 1A |
| chr2_+_113984646 | 0.05 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr8_+_33239139 | 0.05 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr18_-_28017925 | 0.05 |
ENSRNOT00000075420
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr3_+_63379031 | 0.05 |
ENSRNOT00000068199
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr19_-_38484611 | 0.05 |
ENSRNOT00000077960
|
Nfat5
|
nuclear factor of activated T-cells 5 |
| chr4_-_66955732 | 0.05 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr1_-_197568165 | 0.05 |
ENSRNOT00000079044
|
Xpo6
|
exportin 6 |
| chr13_-_86671515 | 0.05 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr5_-_48165317 | 0.04 |
ENSRNOT00000088179
|
Ankrd6
|
ankyrin repeat domain 6 |
| chr14_+_32257805 | 0.04 |
ENSRNOT00000039626
|
AABR07014804.1
|
|
| chr7_+_107406962 | 0.04 |
ENSRNOT00000093254
|
Phf20l1
|
PHD finger protein 20-like protein 1 |
| chr6_-_71199110 | 0.04 |
ENSRNOT00000081883
|
Prkd1
|
protein kinase D1 |
| chr9_+_73418607 | 0.04 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr1_-_164977633 | 0.04 |
ENSRNOT00000029629
|
Rnf169
|
ring finger protein 169 |
| chr1_-_128287151 | 0.04 |
ENSRNOT00000084946
ENSRNOT00000089723 |
Mef2a
|
myocyte enhancer factor 2a |
| chr1_-_126211439 | 0.03 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr19_-_39267928 | 0.03 |
ENSRNOT00000027686
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr17_-_76188979 | 0.03 |
ENSRNOT00000092442
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr2_+_123597340 | 0.03 |
ENSRNOT00000058552
|
RGD1307100
|
similar to RIKEN cDNA D630029K19 |
| chr5_+_116420690 | 0.03 |
ENSRNOT00000087089
|
Nfia
|
nuclear factor I/A |
| chr13_+_98231326 | 0.03 |
ENSRNOT00000003837
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr16_+_39827749 | 0.03 |
ENSRNOT00000074885
|
Wdr17
|
WD repeat domain 17 |
| chr17_-_15467320 | 0.03 |
ENSRNOT00000072490
ENSRNOT00000093561 |
Ogn
|
osteoglycin |
| chr19_-_11669578 | 0.03 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr1_-_101819478 | 0.03 |
ENSRNOT00000056181
|
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr6_+_48452369 | 0.03 |
ENSRNOT00000044310
|
Myt1l
|
myelin transcription factor 1-like |
| chr8_+_82043403 | 0.03 |
ENSRNOT00000091789
|
Myo5a
|
myosin VA |
| chr2_+_55747318 | 0.03 |
ENSRNOT00000050655
|
Dab2
|
DAB2, clathrin adaptor protein |
| chr11_-_4397361 | 0.03 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr7_-_140291620 | 0.02 |
ENSRNOT00000088323
|
Adcy6
|
adenylate cyclase 6 |
| chr17_-_76281660 | 0.02 |
ENSRNOT00000058070
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr9_+_77320726 | 0.02 |
ENSRNOT00000068450
|
Spag16
|
sperm associated antigen 16 |
| chr5_+_156439633 | 0.02 |
ENSRNOT00000090232
|
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr13_-_88436789 | 0.02 |
ENSRNOT00000003901
|
Ddr2
|
discoidin domain receptor tyrosine kinase 2 |
| chr3_+_104816987 | 0.02 |
ENSRNOT00000042103
ENSRNOT00000044625 |
Fmn1
|
formin 1 |
| chr11_-_83300409 | 0.02 |
ENSRNOT00000029255
|
Vps8
|
VPS8 CORVET complex subunit |
| chr7_-_15163866 | 0.02 |
ENSRNOT00000042638
|
Zfp870
|
zinc finger protein 870 |
| chr2_-_88763733 | 0.02 |
ENSRNOT00000059424
|
LOC688389
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr4_+_161720501 | 0.02 |
ENSRNOT00000008196
ENSRNOT00000055886 |
Nrip2
|
nuclear receptor interacting protein 2 |
| chr1_+_88955440 | 0.02 |
ENSRNOT00000091101
|
Prodh2
|
proline dehydrogenase 2 |
| chr1_+_170205591 | 0.02 |
ENSRNOT00000071063
|
LOC686660
|
similar to olfactory receptor 692 |
| chr2_+_209661244 | 0.02 |
ENSRNOT00000091973
|
AABR07012826.1
|
|
| chrM_+_2740 | 0.02 |
ENSRNOT00000047550
|
Mt-nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr5_+_36566783 | 0.02 |
ENSRNOT00000077650
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr11_-_72814850 | 0.01 |
ENSRNOT00000091575
|
LOC100911374
|
UBX domain-containing protein 7-like |
| chr1_-_266428239 | 0.01 |
ENSRNOT00000027160
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr10_+_89285855 | 0.01 |
ENSRNOT00000028033
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr16_-_48692476 | 0.01 |
ENSRNOT00000013118
|
Irf2
|
interferon regulatory factor 2 |
| chr1_-_205030567 | 0.01 |
ENSRNOT00000023404
|
Ctbp2
|
C-terminal binding protein 2 |
| chr1_+_261291870 | 0.01 |
ENSRNOT00000049914
|
Hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr2_-_27287605 | 0.01 |
ENSRNOT00000034041
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr20_-_3605638 | 0.01 |
ENSRNOT00000074460
|
Sfta2
|
surfactant associated 2 |
| chr2_-_157759819 | 0.01 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chrM_+_9451 | 0.01 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chrM_+_14136 | 0.01 |
ENSRNOT00000042098
|
Mt-cyb
|
mitochondrially encoded cytochrome b |
| chr9_+_53013413 | 0.01 |
ENSRNOT00000005313
|
Ankar
|
ankyrin and armadillo repeat containing |
| chr16_+_8211420 | 0.01 |
ENSRNOT00000079609
|
Oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr7_-_12601674 | 0.01 |
ENSRNOT00000093489
|
Arid3a
|
AT-rich interaction domain 3A |
| chr12_+_21981755 | 0.01 |
ENSRNOT00000001871
|
LOC100910540
|
7SK snRNA methylphosphate capping enzyme-like |
| chr13_-_74331214 | 0.01 |
ENSRNOT00000006018
|
Fam20b
|
FAM20B, glycosaminoglycan xylosylkinase |
| chr2_-_88660449 | 0.01 |
ENSRNOT00000051741
|
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr2_-_184263564 | 0.01 |
ENSRNOT00000015279
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr2_+_235264219 | 0.01 |
ENSRNOT00000086245
|
Cfi
|
complement factor I |
| chr1_-_99632845 | 0.01 |
ENSRNOT00000049123
|
LOC103691107
|
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4 pseudogene |
| chr17_-_54714914 | 0.01 |
ENSRNOT00000024336
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr1_+_88955135 | 0.01 |
ENSRNOT00000083550
|
Prodh2
|
proline dehydrogenase 2 |
| chr11_+_65743892 | 0.00 |
ENSRNOT00000085226
|
AC106292.2
|
|
| chr14_-_42560174 | 0.00 |
ENSRNOT00000003128
|
Tmem33
|
transmembrane protein 33 |
| chr5_-_128730446 | 0.00 |
ENSRNOT00000089441
|
Osbpl9
|
oxysterol binding protein-like 9 |
| chr8_-_126390801 | 0.00 |
ENSRNOT00000089732
|
AABR07071661.1
|
|
| chr20_-_4132604 | 0.00 |
ENSRNOT00000077630
ENSRNOT00000048332 |
RT1-Da
|
RT1 class II, locus Da |
| chr17_-_78499881 | 0.00 |
ENSRNOT00000079260
|
Fam107b
|
family with sequence similarity 107, member B |
| chrX_+_54390733 | 0.00 |
ENSRNOT00000004977
|
RGD1565785
|
similar to chromosome X open reading frame 21 |
| chr1_+_107262659 | 0.00 |
ENSRNOT00000022499
|
Gas2
|
growth arrest-specific 2 |
| chr6_-_33738825 | 0.00 |
ENSRNOT00000039492
|
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfhx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0048382 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) mesendoderm development(GO:0048382) regulation of cardiac ventricle development(GO:1904412) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |