Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
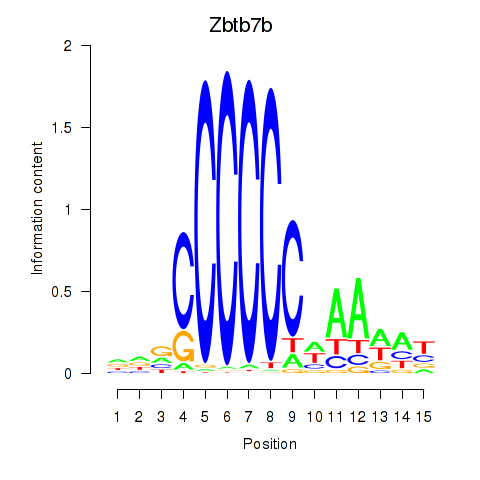
Results for Zbtb7b
Z-value: 0.66
Transcription factors associated with Zbtb7b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7b
|
ENSRNOG00000020640 | zinc finger and BTB domain containing 7B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7b | rn6_v1_chr2_-_188718704_188718704 | 0.09 | 8.9e-01 | Click! |
Activity profile of Zbtb7b motif
Sorted Z-values of Zbtb7b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_168945449 | 0.48 |
ENSRNOT00000087661
ENSRNOT00000019913 |
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr1_-_72339395 | 0.41 |
ENSRNOT00000021772
|
Zfp580
|
zinc finger protein 580 |
| chr14_+_34389991 | 0.38 |
ENSRNOT00000002953
|
Pdcl2
|
phosducin-like 2 |
| chr1_+_168964202 | 0.32 |
ENSRNOT00000089102
|
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr6_-_123577695 | 0.32 |
ENSRNOT00000006604
|
Foxn3
|
forkhead box N3 |
| chr8_-_98738446 | 0.30 |
ENSRNOT00000019860
|
Zic1
|
Zic family member 1 |
| chr5_-_77945016 | 0.27 |
ENSRNOT00000076223
|
Zfp37
|
zinc finger protein 37 |
| chr12_+_18531990 | 0.26 |
ENSRNOT00000036606
|
Asmtl
|
acetylserotonin O-methyltransferase-like |
| chr20_+_14577166 | 0.26 |
ENSRNOT00000085583
|
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr7_-_11018160 | 0.26 |
ENSRNOT00000092061
|
Aes
|
amino-terminal enhancer of split |
| chr1_+_101178104 | 0.24 |
ENSRNOT00000028072
|
Pth2
|
parathyroid hormone 2 |
| chr5_-_136762986 | 0.23 |
ENSRNOT00000026852
|
Artn
|
artemin |
| chr6_+_33885495 | 0.21 |
ENSRNOT00000086633
|
Sdc1
|
syndecan 1 |
| chr15_-_60289763 | 0.19 |
ENSRNOT00000038579
|
Fam216b
|
family with sequence similarity 216, member B |
| chr6_+_51662224 | 0.16 |
ENSRNOT00000060006
|
Ccdc71l
|
coiled-coil domain containing 71-like |
| chr12_+_18516946 | 0.16 |
ENSRNOT00000029485
|
Dhrsx
|
dehydrogenase/reductase X-linked |
| chr1_-_255376833 | 0.15 |
ENSRNOT00000024941
|
Ppp1r3c
|
protein phosphatase 1, regulatory subunit 3C |
| chr6_-_111267734 | 0.15 |
ENSRNOT00000074037
|
Noxred1
|
NADP-dependent oxidoreductase domain containing 1 |
| chr15_+_33600102 | 0.15 |
ENSRNOT00000022664
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chrX_-_1369384 | 0.14 |
ENSRNOT00000013745
|
Timp1
|
TIMP metallopeptidase inhibitor 1 |
| chr15_+_33600337 | 0.14 |
ENSRNOT00000075965
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr19_-_36157924 | 0.14 |
ENSRNOT00000072022
|
AABR07043701.1
|
|
| chr18_-_15540177 | 0.14 |
ENSRNOT00000022113
|
Ttr
|
transthyretin |
| chr19_-_37427989 | 0.13 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr9_-_9912276 | 0.13 |
ENSRNOT00000071515
|
Tnfsf9
|
tumor necrosis factor superfamily member 9 |
| chr6_-_104409005 | 0.13 |
ENSRNOT00000040965
|
Ccdc177
|
coiled-coil domain containing 177 |
| chr19_-_55257876 | 0.12 |
ENSRNOT00000017564
|
Cyba
|
cytochrome b-245 alpha chain |
| chr6_+_129538982 | 0.12 |
ENSRNOT00000083626
ENSRNOT00000082522 |
Ak7
|
adenylate kinase 7 |
| chr18_-_5314511 | 0.12 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr1_-_164307084 | 0.12 |
ENSRNOT00000086091
|
Serpinh1
|
serpin family H member 1 |
| chr12_+_2046472 | 0.12 |
ENSRNOT00000001289
|
Zfp358
|
zinc finger protein 358 |
| chr11_-_87081950 | 0.11 |
ENSRNOT00000002574
|
Dgcr6
|
DiGeorge syndrome critical region gene 6 |
| chr20_+_4823662 | 0.11 |
ENSRNOT00000090028
|
Nfkbil1
|
NFKB inhibitor like 1 |
| chr10_+_66099531 | 0.11 |
ENSRNOT00000056192
|
Lyrm9
|
LYR motif containing 9 |
| chr20_+_5456235 | 0.11 |
ENSRNOT00000092555
|
Pfdn6
|
prefoldin subunit 6 |
| chr18_-_12640716 | 0.11 |
ENSRNOT00000020697
|
Klhl14
|
kelch-like family member 14 |
| chr20_+_4824226 | 0.10 |
ENSRNOT00000001113
|
Nfkbil1
|
NFKB inhibitor like 1 |
| chr7_+_12820840 | 0.09 |
ENSRNOT00000012317
|
Rnf126
|
ring finger protein 126 |
| chr1_-_80185882 | 0.09 |
ENSRNOT00000022214
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr1_+_64101018 | 0.09 |
ENSRNOT00000078640
|
Mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr10_+_72486152 | 0.09 |
ENSRNOT00000004193
|
RGD1310166
|
similar to Chromodomain-helicase-DNA-binding protein 1 (CHD-1) |
| chr10_+_103438303 | 0.08 |
ENSRNOT00000085262
|
Cd300a
|
Cd300a molecule |
| chr18_+_32273770 | 0.08 |
ENSRNOT00000087408
|
Fgf1
|
fibroblast growth factor 1 |
| chr2_-_198719202 | 0.08 |
ENSRNOT00000028801
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr2_+_225645568 | 0.08 |
ENSRNOT00000017878
|
Abca4
|
ATP binding cassette subfamily A member 4 |
| chr20_-_4380600 | 0.08 |
ENSRNOT00000068712
|
Egfl8
|
EGF-like-domain, multiple 8 |
| chr6_+_42854888 | 0.08 |
ENSRNOT00000007259
|
Odc1
|
ornithine decarboxylase 1 |
| chr15_+_57505449 | 0.08 |
ENSRNOT00000064945
|
Siah3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr8_-_22785671 | 0.08 |
ENSRNOT00000045384
|
Spc24
|
SPC24, NDC80 kinetochore complex component |
| chr12_-_42492526 | 0.08 |
ENSRNOT00000084018
|
Tbx3
|
T-box 3 |
| chr1_-_167347490 | 0.08 |
ENSRNOT00000076499
|
Rhog
|
ras homolog family member G |
| chr5_+_159845774 | 0.08 |
ENSRNOT00000012328
|
Epha2
|
Eph receptor A2 |
| chr8_-_46694613 | 0.07 |
ENSRNOT00000084514
|
AC133265.1
|
|
| chr4_-_55740627 | 0.07 |
ENSRNOT00000010658
|
Pax4
|
paired box 4 |
| chr19_+_387374 | 0.07 |
ENSRNOT00000075433
|
LOC102547811
|
uncharacterized LOC102547811 |
| chr1_-_86948845 | 0.07 |
ENSRNOT00000027212
|
Nfkbib
|
NFKB inhibitor beta |
| chr7_-_11478489 | 0.07 |
ENSRNOT00000027272
|
Map2k2
|
mitogen activated protein kinase kinase 2 |
| chr3_+_95711555 | 0.07 |
ENSRNOT00000006302
|
Pax6
|
paired box 6 |
| chr18_-_19275273 | 0.07 |
ENSRNOT00000041707
|
LOC102547344
|
lateral signaling target protein 2 homolog |
| chr5_-_2982603 | 0.07 |
ENSRNOT00000045460
|
Kcnb2
|
potassium voltage-gated channel subfamily B member 2 |
| chr9_-_16612136 | 0.07 |
ENSRNOT00000023495
|
Mea1
|
male-enhanced antigen 1 |
| chr10_+_106812739 | 0.07 |
ENSRNOT00000074225
|
Syngr2
|
synaptogyrin 2 |
| chr7_+_121311024 | 0.06 |
ENSRNOT00000092260
ENSRNOT00000023066 ENSRNOT00000081377 |
Syngr1
|
synaptogyrin 1 |
| chr20_-_29579578 | 0.06 |
ENSRNOT00000000700
|
AC096404.1
|
|
| chr20_-_4823475 | 0.06 |
ENSRNOT00000082536
ENSRNOT00000001114 |
Atp6v1g2
|
ATPase H+ transporting V1 subunit G2 |
| chr2_+_30685840 | 0.06 |
ENSRNOT00000031385
|
Ccdc125
|
coiled-coil domain containing 125 |
| chr5_-_139748489 | 0.06 |
ENSRNOT00000078741
|
Nfyc
|
nuclear transcription factor Y subunit gamma |
| chr1_-_65659945 | 0.05 |
ENSRNOT00000058574
|
Rnf225
|
ring finger protein 225 |
| chr20_+_4392626 | 0.05 |
ENSRNOT00000000495
|
Prrt1
|
proline-rich transmembrane protein 1 |
| chr1_-_80615704 | 0.05 |
ENSRNOT00000041891
|
Apoe
|
apolipoprotein E |
| chr6_+_126018841 | 0.05 |
ENSRNOT00000089840
ENSRNOT00000088089 |
Slc24a4
|
solute carrier family 24 member 4 |
| chr7_-_123101851 | 0.05 |
ENSRNOT00000090984
ENSRNOT00000005837 |
Phf5a
|
PHD finger protein 5A |
| chr10_+_105235289 | 0.05 |
ENSRNOT00000075548
|
Galr2
|
galanin receptor 2 |
| chr2_-_198120041 | 0.05 |
ENSRNOT00000079621
ENSRNOT00000028747 |
Plekho1
|
pleckstrin homology domain containing O1 |
| chr12_-_23624212 | 0.05 |
ENSRNOT00000064405
ENSRNOT00000001943 |
Rasa4
|
RAS p21 protein activator 4 |
| chr1_+_72580424 | 0.05 |
ENSRNOT00000022977
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr10_+_105156632 | 0.05 |
ENSRNOT00000086288
|
Galr2
|
galanin receptor 2 |
| chr20_-_4392343 | 0.05 |
ENSRNOT00000080476
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr20_+_13778178 | 0.05 |
ENSRNOT00000058314
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr19_+_26084903 | 0.05 |
ENSRNOT00000004799
|
Prdx2
|
peroxiredoxin 2 |
| chr15_+_33755478 | 0.05 |
ENSRNOT00000034073
|
Thtpa
|
thiamine triphosphatase |
| chr9_+_3896337 | 0.05 |
ENSRNOT00000079166
|
LOC103693189
|
protein tyrosine phosphatase type IVA 1 |
| chr12_+_22229079 | 0.05 |
ENSRNOT00000001911
|
Gnb2
|
G protein subunit beta 2 |
| chr10_+_96661696 | 0.05 |
ENSRNOT00000077567
|
AABR07030630.1
|
|
| chr4_+_118852062 | 0.05 |
ENSRNOT00000090827
ENSRNOT00000025070 |
Gfpt1
|
glutamine fructose-6-phosphate transaminase 1 |
| chr1_-_89488223 | 0.05 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr1_+_79898551 | 0.05 |
ENSRNOT00000019069
|
Mypop
|
Myb-related transcription factor, partner of profilin |
| chr7_-_11777503 | 0.04 |
ENSRNOT00000026220
|
Amh
|
anti-Mullerian hormone |
| chr1_+_53174879 | 0.04 |
ENSRNOT00000017601
ENSRNOT00000084628 |
Rnaset2
|
ribonuclease T2 |
| chr1_-_80615525 | 0.04 |
ENSRNOT00000091574
|
Apoe
|
apolipoprotein E |
| chr20_-_5020150 | 0.04 |
ENSRNOT00000001146
|
Sapcd1
|
suppressor APC domain containing 1 |
| chr12_-_22417980 | 0.04 |
ENSRNOT00000072208
|
Ephb4
|
EPH receptor B4 |
| chr1_-_101085884 | 0.04 |
ENSRNOT00000085352
|
Rcn3
|
reticulocalbin 3 |
| chr11_-_61234944 | 0.04 |
ENSRNOT00000059680
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr1_-_221431713 | 0.04 |
ENSRNOT00000028485
|
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr5_-_78393143 | 0.04 |
ENSRNOT00000043610
|
LOC100910017
|
60S ribosomal protein L31-like |
| chr10_-_95434688 | 0.04 |
ENSRNOT00000074809
|
AABR07030603.1
|
|
| chr4_+_98027554 | 0.04 |
ENSRNOT00000009503
|
Serbp1
|
Serpine1 mRNA binding protein 1 |
| chr1_-_185569190 | 0.04 |
ENSRNOT00000090773
|
RGD1311703
|
similar to sid2057p |
| chr7_-_143473697 | 0.04 |
ENSRNOT00000055336
ENSRNOT00000083027 |
Krt77
|
keratin 77 |
| chr12_-_42492328 | 0.04 |
ENSRNOT00000011552
|
Tbx3
|
T-box 3 |
| chr15_-_51386190 | 0.04 |
ENSRNOT00000022706
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr14_+_113968563 | 0.04 |
ENSRNOT00000006085
|
Clhc1
|
clathrin heavy chain linker domain containing 1 |
| chr1_+_86948918 | 0.04 |
ENSRNOT00000084839
|
Sirt2
|
sirtuin 2 |
| chr3_-_176459533 | 0.04 |
ENSRNOT00000079283
|
Nkain4
|
Sodium/potassium transporting ATPase interacting 4 |
| chr13_-_50535110 | 0.04 |
ENSRNOT00000077054
|
Kiss1
|
KiSS-1 metastasis-suppressor |
| chr10_-_15311189 | 0.04 |
ENSRNOT00000027371
|
Prr35
|
proline rich 35 |
| chr20_-_11340296 | 0.04 |
ENSRNOT00000038547
|
Icoslg
|
inducible T-cell co-stimulator ligand |
| chr1_+_100593892 | 0.04 |
ENSRNOT00000027062
|
Kcnc3
|
potassium voltage-gated channel subfamily C member 3 |
| chr19_+_60104804 | 0.04 |
ENSRNOT00000078559
|
Pard3
|
par-3 family cell polarity regulator |
| chr11_-_89260297 | 0.04 |
ENSRNOT00000057502
|
Spidr
|
scaffolding protein involved in DNA repair |
| chr11_+_30363280 | 0.04 |
ENSRNOT00000002885
|
Sod1
|
superoxide dismutase 1, soluble |
| chr9_+_82436458 | 0.04 |
ENSRNOT00000026467
ENSRNOT00000081958 |
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr20_+_4855829 | 0.03 |
ENSRNOT00000001110
|
LOC103694380
|
tumor necrosis factor-like |
| chr8_-_33661049 | 0.03 |
ENSRNOT00000068037
|
Fli1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr20_+_20577561 | 0.03 |
ENSRNOT00000081113
|
Cdk1
|
cyclin-dependent kinase 1 |
| chr1_-_225329770 | 0.03 |
ENSRNOT00000027459
|
Asrgl1
|
asparaginase like 1 |
| chr6_+_12362813 | 0.03 |
ENSRNOT00000022370
|
Ston1
|
stonin 1 |
| chr4_-_179307095 | 0.03 |
ENSRNOT00000021193
|
Bcat1
|
branched chain amino acid transaminase 1 |
| chr8_+_64325435 | 0.03 |
ENSRNOT00000013747
|
Hexa
|
hexosaminidase subunit alpha |
| chr18_+_76559811 | 0.03 |
ENSRNOT00000084621
|
Pard6g
|
par-6 family cell polarity regulator gamma |
| chr10_+_82800704 | 0.03 |
ENSRNOT00000089497
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chr1_-_71399751 | 0.03 |
ENSRNOT00000020787
|
Zfp444
|
zinc finger protein 444 |
| chr3_+_62481323 | 0.03 |
ENSRNOT00000078872
|
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr6_+_107428508 | 0.03 |
ENSRNOT00000013431
|
RGD1307704
|
similar to RIKEN cDNA 2410016O06 |
| chr1_+_100830064 | 0.03 |
ENSRNOT00000027320
|
Il4i1
|
interleukin 4 induced 1 |
| chr7_+_38900593 | 0.03 |
ENSRNOT00000072166
|
Epyc
|
epiphycan |
| chr16_-_19669153 | 0.03 |
ENSRNOT00000078714
ENSRNOT00000090775 |
Haus8
|
HAUS augmin-like complex, subunit 8 |
| chrX_+_111122552 | 0.03 |
ENSRNOT00000083566
ENSRNOT00000085078 ENSRNOT00000090928 |
Cldn2
|
claudin 2 |
| chr2_+_187951344 | 0.03 |
ENSRNOT00000027123
|
Ssr2
|
signal sequence receptor, beta |
| chr10_+_69737328 | 0.03 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr17_+_70684134 | 0.03 |
ENSRNOT00000025731
ENSRNOT00000068354 |
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr1_+_47972399 | 0.03 |
ENSRNOT00000033408
ENSRNOT00000077707 |
Acat2
|
acetyl-CoA acetyltransferase 2 |
| chr4_-_4473307 | 0.03 |
ENSRNOT00000045773
|
Dpp6
|
dipeptidyl peptidase like 6 |
| chr10_-_14072230 | 0.03 |
ENSRNOT00000018405
|
Tbl3
|
transducin (beta)-like 3 |
| chr13_+_82438697 | 0.03 |
ENSRNOT00000003759
|
Selp
|
selectin P |
| chr12_+_39423596 | 0.03 |
ENSRNOT00000066952
|
Ift81
|
intraflagellar transport 81 |
| chr4_-_157274755 | 0.03 |
ENSRNOT00000088895
|
LOC100911672
|
atrophin-1-like |
| chr20_+_5455974 | 0.03 |
ENSRNOT00000000553
ENSRNOT00000092676 |
Pfdn6
|
prefoldin subunit 6 |
| chr9_+_17216495 | 0.03 |
ENSRNOT00000026331
|
Mad2l1bp
|
MAD2L1 binding protein |
| chr7_+_116671948 | 0.02 |
ENSRNOT00000077773
ENSRNOT00000029711 |
Gli4
|
GLI family zinc finger 4 |
| chr12_-_11175917 | 0.02 |
ENSRNOT00000079573
|
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr16_-_19872774 | 0.02 |
ENSRNOT00000000060
|
Abhd8
|
abhydrolase domain containing 8 |
| chr10_+_86819472 | 0.02 |
ENSRNOT00000081424
|
Cdc6
|
cell division cycle 6 |
| chr13_+_48689962 | 0.02 |
ENSRNOT00000079707
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr8_-_85645718 | 0.02 |
ENSRNOT00000032185
|
Gsta2
|
glutathione S-transferase alpha 2 |
| chrX_+_77065397 | 0.02 |
ENSRNOT00000090007
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr17_-_42031265 | 0.02 |
ENSRNOT00000068021
|
Dcdc2
|
doublecortin domain containing 2 |
| chr18_-_80865584 | 0.02 |
ENSRNOT00000021750
|
Tshz1
|
teashirt zinc finger homeobox 1 |
| chr1_-_88780425 | 0.02 |
ENSRNOT00000074494
|
Sdhaf1
|
succinate dehydrogenase complex assembly factor 1 |
| chr19_+_25095089 | 0.02 |
ENSRNOT00000041717
|
Prkaca
|
protein kinase cAMP-activated catalytic subunit alpha |
| chr9_+_16612433 | 0.02 |
ENSRNOT00000023979
|
Klhdc3
|
kelch domain containing 3 |
| chr12_-_30180115 | 0.02 |
ENSRNOT00000001202
|
Crcp
|
CGRP receptor component |
| chr1_-_220974597 | 0.02 |
ENSRNOT00000088799
|
Kat5
|
lysine acetyltransferase 5 |
| chr16_-_20641908 | 0.02 |
ENSRNOT00000026846
|
Ell
|
elongation factor for RNA polymerase II |
| chr3_-_107760550 | 0.02 |
ENSRNOT00000077091
ENSRNOT00000051638 |
Meis2
|
Meis homeobox 2 |
| chr1_+_214434626 | 0.02 |
ENSRNOT00000025319
|
Pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr20_-_2211995 | 0.02 |
ENSRNOT00000077201
|
Trim26
|
tripartite motif-containing 26 |
| chr15_-_28313841 | 0.02 |
ENSRNOT00000085897
|
Ndrg2
|
NDRG family member 2 |
| chr8_+_50310405 | 0.02 |
ENSRNOT00000073507
|
Sik3
|
SIK family kinase 3 |
| chr10_-_86004096 | 0.02 |
ENSRNOT00000091978
ENSRNOT00000066855 |
Stac2
|
SH3 and cysteine rich domain 2 |
| chr20_-_10257044 | 0.02 |
ENSRNOT00000068289
|
Wdr4
|
WD repeat domain 4 |
| chr12_+_4248808 | 0.02 |
ENSRNOT00000042410
|
AABR07035089.1
|
|
| chr13_+_60545635 | 0.02 |
ENSRNOT00000077153
|
Uchl5
|
ubiquitin C-terminal hydrolase L5 |
| chr3_-_160573978 | 0.02 |
ENSRNOT00000018386
|
Wfdc5
|
WAP four-disulfide core domain 5 |
| chr10_-_14703668 | 0.01 |
ENSRNOT00000024767
|
Tpsab1
|
tryptase alpha/beta 1 |
| chr2_+_189430041 | 0.01 |
ENSRNOT00000023567
ENSRNOT00000023605 |
Tpm3
|
tropomyosin 3 |
| chr3_-_160465143 | 0.01 |
ENSRNOT00000040156
|
Tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr3_-_113023532 | 0.01 |
ENSRNOT00000084921
|
Tgm7l1
|
transglutaminase 7-like 1 |
| chr14_-_113968268 | 0.01 |
ENSRNOT00000005872
|
Rps27a
|
ribosomal protein S27a |
| chr3_+_147422095 | 0.01 |
ENSRNOT00000006786
|
Angpt4
|
angiopoietin 4 |
| chr11_+_45751812 | 0.01 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr16_-_81583916 | 0.01 |
ENSRNOT00000026281
|
LOC103693999
|
transmembrane and coiled-coil domain-containing protein 3 |
| chr3_-_48535909 | 0.01 |
ENSRNOT00000008148
|
Fap
|
fibroblast activation protein, alpha |
| chr20_-_5455632 | 0.01 |
ENSRNOT00000000552
|
Wdr46
|
WD repeat domain 46 |
| chr3_+_119677575 | 0.01 |
ENSRNOT00000031577
|
Tmem127
|
transmembrane protein 127 |
| chrX_-_13279082 | 0.01 |
ENSRNOT00000051898
ENSRNOT00000060857 |
Tspan7
|
tetraspanin 7 |
| chr3_-_44522930 | 0.01 |
ENSRNOT00000006963
|
Acvr1
|
activin A receptor type 1 |
| chr5_+_154868534 | 0.01 |
ENSRNOT00000091442
|
Luzp1
|
leucine zipper protein 1 |
| chr1_-_80599572 | 0.01 |
ENSRNOT00000024832
|
Apoc4
|
apolipoprotein C4 |
| chr5_+_57472315 | 0.01 |
ENSRNOT00000015575
|
Ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr2_+_60180215 | 0.01 |
ENSRNOT00000084624
|
Prlr
|
prolactin receptor |
| chr2_-_184951950 | 0.01 |
ENSRNOT00000093486
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr11_+_86797557 | 0.01 |
ENSRNOT00000083049
ENSRNOT00000046594 |
Tango2
|
transport and golgi organization 2 homolog |
| chr13_-_89668473 | 0.01 |
ENSRNOT00000004938
|
Ufc1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr14_-_79987915 | 0.01 |
ENSRNOT00000089006
|
Afap1
|
actin filament associated protein 1 |
| chr17_-_32953641 | 0.01 |
ENSRNOT00000023332
|
Wrnip1
|
Werner helicase interacting protein 1 |
| chr4_-_66624912 | 0.01 |
ENSRNOT00000064891
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr20_-_7930929 | 0.01 |
ENSRNOT00000000607
|
Tead3
|
TEA domain transcription factor 3 |
| chr16_-_20864176 | 0.01 |
ENSRNOT00000092706
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr13_+_106751625 | 0.01 |
ENSRNOT00000004992
|
Ush2a
|
usherin |
| chr1_-_82409639 | 0.01 |
ENSRNOT00000031326
|
Erich4
|
glutamate-rich 4 |
| chr4_-_82160240 | 0.01 |
ENSRNOT00000038775
ENSRNOT00000058985 |
Hoxa4
|
homeo box A4 |
| chr9_-_76768770 | 0.01 |
ENSRNOT00000087779
ENSRNOT00000057849 |
Ikzf2
|
IKAROS family zinc finger 2 |
| chr15_-_5894854 | 0.01 |
ENSRNOT00000024427
|
Cd99l2
|
CD99 molecule-like 2 |
| chr2_+_186630278 | 0.01 |
ENSRNOT00000021604
|
LOC365839
|
similar to elongation protein 4 homolog |
| chr1_-_217721376 | 0.01 |
ENSRNOT00000092748
|
Ppfia1
|
PTPRF interacting protein alpha 1 |
| chr3_-_154490851 | 0.00 |
ENSRNOT00000017213
|
Tti1
|
TELO2 interacting protein 1 |
| chr7_-_2623781 | 0.00 |
ENSRNOT00000004173
|
Spryd4
|
SPRY domain containing 4 |
| chr1_+_243589607 | 0.00 |
ENSRNOT00000021792
|
Dmrt3
|
doublesex and mab-3 related transcription factor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 0.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.4 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:1903002 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.3 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:2000417 | negative regulation of phagocytosis, engulfment(GO:0060101) negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.0 | 0.1 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:1904937 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.0 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.1 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.0 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:1901073 | glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.0 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.0 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.0 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.0 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.0 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |