Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
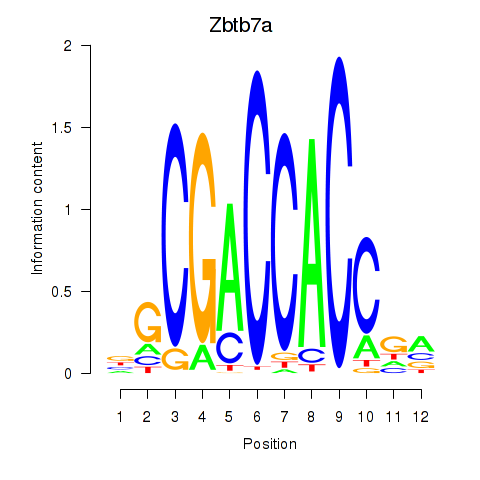
Results for Zbtb7a
Z-value: 0.36
Transcription factors associated with Zbtb7a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7a
|
ENSRNOG00000020161 | zinc finger and BTB domain containing 7a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7a | rn6_v1_chr7_-_11444786_11444786 | 0.59 | 2.9e-01 | Click! |
Activity profile of Zbtb7a motif
Sorted Z-values of Zbtb7a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_25839198 | 0.12 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr1_+_91363492 | 0.09 |
ENSRNOT00000014517
|
Cebpa
|
CCAAT/enhancer binding protein alpha |
| chr17_-_21353134 | 0.08 |
ENSRNOT00000067898
|
Smim13
|
small integral membrane protein 13 |
| chr10_+_35279236 | 0.08 |
ENSRNOT00000003770
|
Gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chrX_+_76786466 | 0.08 |
ENSRNOT00000090665
|
Fgf16
|
fibroblast growth factor 16 |
| chr19_+_23389375 | 0.08 |
ENSRNOT00000018629
|
Sall1
|
spalt-like transcription factor 1 |
| chr5_-_151397603 | 0.07 |
ENSRNOT00000076866
|
Gpr3
|
G protein-coupled receptor 3 |
| chr4_-_157433467 | 0.07 |
ENSRNOT00000028965
|
Lag3
|
lymphocyte activating 3 |
| chr18_-_28454756 | 0.07 |
ENSRNOT00000040091
|
Spata24
|
spermatogenesis associated 24 |
| chr16_+_80826681 | 0.07 |
ENSRNOT00000000123
|
Coprs
|
coordinator of PRMT5 and differentiation stimulator |
| chr18_+_29110242 | 0.07 |
ENSRNOT00000025756
|
Pura
|
purine rich element binding protein A |
| chr5_-_135677432 | 0.07 |
ENSRNOT00000024393
|
Hpdl
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr1_-_101175570 | 0.07 |
ENSRNOT00000073918
|
Gfy
|
golgi-associated, olfactory signaling regulator |
| chr5_+_152681101 | 0.07 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr1_+_256382791 | 0.07 |
ENSRNOT00000022549
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr17_-_78910671 | 0.06 |
ENSRNOT00000064609
|
Acbd7
|
acyl-CoA binding domain containing 7 |
| chr16_-_62483137 | 0.06 |
ENSRNOT00000020683
|
Purg
|
purine-rich element binding protein G |
| chr6_+_104718512 | 0.06 |
ENSRNOT00000007947
|
Smoc1
|
SPARC related modular calcium binding 1 |
| chr10_+_16970626 | 0.06 |
ENSRNOT00000005383
|
Dusp1
|
dual specificity phosphatase 1 |
| chr1_+_32058044 | 0.06 |
ENSRNOT00000021898
|
Nkd2
|
naked cuticle homolog 2 |
| chr12_-_30033357 | 0.06 |
ENSRNOT00000001198
|
Kctd7
|
potassium channel tetramerization domain containing 7 |
| chr3_+_175885894 | 0.06 |
ENSRNOT00000039780
|
Ntsr1
|
neurotensin receptor 1 |
| chr10_-_96584896 | 0.06 |
ENSRNOT00000004699
ENSRNOT00000055073 |
Prkca
|
protein kinase C, alpha |
| chr12_-_17972737 | 0.05 |
ENSRNOT00000001783
|
Fam20c
|
FAM20C, golgi associated secretory pathway kinase |
| chr7_-_12793711 | 0.05 |
ENSRNOT00000013762
|
Palm
|
paralemmin |
| chr4_-_158705885 | 0.05 |
ENSRNOT00000087115
ENSRNOT00000026685 |
Ntf3
|
neurotrophin 3 |
| chr4_+_148199102 | 0.05 |
ENSRNOT00000083563
|
Zfand4
|
zinc finger AN1-type containing 4 |
| chr1_+_104576589 | 0.05 |
ENSRNOT00000046529
|
Nav2
|
neuron navigator 2 |
| chr4_-_100660140 | 0.05 |
ENSRNOT00000020005
|
Tcf7l1
|
transcription factor 7 like 1 |
| chr15_+_12827707 | 0.05 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr3_+_22640545 | 0.05 |
ENSRNOT00000064507
ENSRNOT00000014452 |
Lhx2
|
LIM homeobox 2 |
| chr3_-_165741967 | 0.05 |
ENSRNOT00000017063
|
Zfp64
|
zinc finger protein 64 |
| chr5_-_36683356 | 0.05 |
ENSRNOT00000009043
|
Pou3f2
|
POU class 3 homeobox 2 |
| chr1_-_239057732 | 0.05 |
ENSRNOT00000024775
|
Gda
|
guanine deaminase |
| chr14_-_18849258 | 0.05 |
ENSRNOT00000033406
|
Pf4
|
platelet factor 4 |
| chr1_-_163328591 | 0.05 |
ENSRNOT00000034843
|
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr3_+_160231914 | 0.05 |
ENSRNOT00000014411
|
Kcnk15
|
potassium two pore domain channel subfamily K member 15 |
| chr1_-_153740905 | 0.05 |
ENSRNOT00000023239
|
Prss23
|
protease, serine, 23 |
| chr9_+_12740885 | 0.04 |
ENSRNOT00000015210
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr8_-_50526843 | 0.04 |
ENSRNOT00000092188
|
AABR07070085.1
|
|
| chr4_-_117631716 | 0.04 |
ENSRNOT00000057475
|
LOC103690006
|
RNA/RNP complex-1-interacting phosphatase |
| chr3_+_7422820 | 0.04 |
ENSRNOT00000064323
|
Ddx31
|
DEAD-box helicase 31 |
| chr6_+_26836216 | 0.04 |
ENSRNOT00000077903
ENSRNOT00000011373 |
Ost4
|
oligosaccharyltransferase complex subunit 4, non-catalytic |
| chr10_-_96015499 | 0.04 |
ENSRNOT00000004383
|
Cacng4
|
calcium voltage-gated channel auxiliary subunit gamma 4 |
| chr2_-_119140110 | 0.04 |
ENSRNOT00000058810
|
AABR07009978.1
|
|
| chr7_-_77162148 | 0.04 |
ENSRNOT00000008350
|
Klf10
|
Kruppel-like factor 10 |
| chr19_-_838099 | 0.04 |
ENSRNOT00000014234
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr14_+_35683442 | 0.04 |
ENSRNOT00000003085
|
Chic2
|
cysteine-rich hydrophobic domain 2 |
| chr17_-_78962476 | 0.04 |
ENSRNOT00000030228
|
Nmt2
|
N-myristoyltransferase 2 |
| chr12_-_37425596 | 0.04 |
ENSRNOT00000084553
|
Tctn2
|
tectonic family member 2 |
| chr10_+_49259194 | 0.04 |
ENSRNOT00000091100
ENSRNOT00000004390 |
Fbxw10
|
F-box and WD repeat domain containing 10 |
| chr7_-_24668250 | 0.04 |
ENSRNOT00000083806
|
Tmem263
|
transmembrane protein 263 |
| chr10_+_65448950 | 0.04 |
ENSRNOT00000082348
ENSRNOT00000037016 |
Rab34
|
RAB34, member RAS oncogene family |
| chr10_+_94207336 | 0.04 |
ENSRNOT00000010816
|
Kcnh6
|
potassium voltage-gated channel subfamily H member 6 |
| chr7_-_118933812 | 0.04 |
ENSRNOT00000031951
|
Apol9a
|
apolipoprotein L 9a |
| chr3_-_72447801 | 0.04 |
ENSRNOT00000088815
|
P2rx3
|
purinergic receptor P2X 3 |
| chr7_-_12899004 | 0.04 |
ENSRNOT00000011086
|
Gzmm
|
granzyme M |
| chr3_+_175982777 | 0.04 |
ENSRNOT00000082073
|
Ntsr1
|
neurotensin receptor 1 |
| chr17_-_14058642 | 0.04 |
ENSRNOT00000019489
|
Nxnl2
|
nucleoredoxin-like 2 |
| chr8_+_49676540 | 0.04 |
ENSRNOT00000022032
ENSRNOT00000082205 |
Fxyd6
|
FXYD domain-containing ion transport regulator 6 |
| chr1_-_218920094 | 0.04 |
ENSRNOT00000022213
|
Lrp5
|
LDL receptor related protein 5 |
| chr12_+_22165486 | 0.04 |
ENSRNOT00000001890
|
Mospd3
|
motile sperm domain containing 3 |
| chr19_-_41433346 | 0.04 |
ENSRNOT00000022952
|
Cmtr2
|
cap methyltransferase 2 |
| chr3_-_7632345 | 0.04 |
ENSRNOT00000049889
|
Cfap77
|
cilia and flagella associated protein 77 |
| chr2_+_119139717 | 0.04 |
ENSRNOT00000016051
|
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr1_-_179010257 | 0.03 |
ENSRNOT00000017199
|
Rras2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr9_-_116222374 | 0.03 |
ENSRNOT00000090111
ENSRNOT00000067900 |
Arhgap28
|
Rho GTPase activating protein 28 |
| chr19_+_25969255 | 0.03 |
ENSRNOT00000004370
|
Farsa
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chrX_+_156873849 | 0.03 |
ENSRNOT00000085410
|
Arhgap4
|
Rho GTPase activating protein 4 |
| chr3_-_138462063 | 0.03 |
ENSRNOT00000065553
|
Ovol2
|
ovo-like zinc finger 2 |
| chr2_+_30346069 | 0.03 |
ENSRNOT00000076577
|
Serf1
|
small EDRK-rich factor 1 |
| chr1_-_259106948 | 0.03 |
ENSRNOT00000074429
|
LOC100912537
|
tectonic-3-like |
| chr11_-_83546674 | 0.03 |
ENSRNOT00000044896
|
Ephb3
|
Eph receptor B3 |
| chr18_-_325374 | 0.03 |
ENSRNOT00000078032
ENSRNOT00000003638 |
Mtmr1
|
myotubularin related protein 1 |
| chr9_+_93326283 | 0.03 |
ENSRNOT00000024578
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr12_-_46920952 | 0.03 |
ENSRNOT00000001532
|
Msi1
|
musashi RNA-binding protein 1 |
| chr2_-_220535751 | 0.03 |
ENSRNOT00000089082
|
Palmd
|
palmdelphin |
| chr16_+_83824430 | 0.03 |
ENSRNOT00000032918
|
Irs2
|
insulin receptor substrate 2 |
| chr8_-_116532169 | 0.03 |
ENSRNOT00000085364
|
Rbm5
|
RNA binding motif protein 5 |
| chr10_+_15672382 | 0.03 |
ENSRNOT00000027965
|
Rhbdf1
|
rhomboid 5 homolog 1 |
| chr8_-_116531784 | 0.03 |
ENSRNOT00000024529
|
Rbm5
|
RNA binding motif protein 5 |
| chr10_+_4957326 | 0.03 |
ENSRNOT00000003458
|
Socs1
|
suppressor of cytokine signaling 1 |
| chr2_+_209766512 | 0.03 |
ENSRNOT00000092240
|
Kcna3
|
potassium voltage-gated channel subfamily A member 3 |
| chr2_-_148838441 | 0.03 |
ENSRNOT00000018294
|
Erich6
|
glutamate-rich 6 |
| chr2_-_56679955 | 0.03 |
ENSRNOT00000016722
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr8_-_72766389 | 0.03 |
ENSRNOT00000024452
|
Lactb
|
lactamase, beta |
| chr14_+_60657686 | 0.03 |
ENSRNOT00000070892
|
Sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr4_-_82702429 | 0.03 |
ENSRNOT00000011069
|
Hibadh
|
3-hydroxyisobutyrate dehydrogenase |
| chr16_-_20860767 | 0.03 |
ENSRNOT00000027275
ENSRNOT00000092417 |
Gdf1
Cers1
|
growth differentiation factor 1 ceramide synthase 1 |
| chr1_-_259691742 | 0.03 |
ENSRNOT00000088065
|
Tctn3
|
tectonic family member 3 |
| chr3_+_154043873 | 0.03 |
ENSRNOT00000072502
ENSRNOT00000034166 |
Nnat
|
neuronatin |
| chr8_+_102304095 | 0.03 |
ENSRNOT00000011358
|
Slc9a9
|
solute carrier family 9 member A9 |
| chr17_-_79676499 | 0.03 |
ENSRNOT00000022711
|
Itga8
|
integrin subunit alpha 8 |
| chr7_+_12192088 | 0.03 |
ENSRNOT00000040170
|
Mex3d
|
mex-3 RNA binding family member D |
| chr3_+_58084606 | 0.03 |
ENSRNOT00000084797
|
Metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr7_+_120125633 | 0.03 |
ENSRNOT00000012480
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr10_+_45089464 | 0.03 |
ENSRNOT00000073792
|
Map2k3
|
mitogen activated protein kinase kinase 3 |
| chr17_+_83221827 | 0.03 |
ENSRNOT00000000155
|
Plxdc2
|
plexin domain containing 2 |
| chr20_+_48335540 | 0.03 |
ENSRNOT00000000352
|
Cd24
|
CD24 molecule |
| chr4_+_32373641 | 0.03 |
ENSRNOT00000076086
|
Dlx6
|
distal-less homeobox 6 |
| chr1_-_222628596 | 0.03 |
ENSRNOT00000083157
ENSRNOT00000034477 |
RGD1308106
|
LOC361719 |
| chr1_+_198192773 | 0.03 |
ENSRNOT00000087625
|
Mapk3
|
mitogen activated protein kinase 3 |
| chr7_+_13039781 | 0.03 |
ENSRNOT00000067771
ENSRNOT00000090048 |
Mier2
|
mesoderm induction early response 1, family member 2 |
| chr7_-_124491004 | 0.03 |
ENSRNOT00000037710
|
Ttll12
|
tubulin tyrosine ligase like 12 |
| chr14_-_43584343 | 0.03 |
ENSRNOT00000039835
|
Nsun7
|
NOP2/Sun RNA methyltransferase family member 7 |
| chr12_-_37574750 | 0.03 |
ENSRNOT00000066253
|
Kmt5a
|
lysine methyltransferase 5A |
| chr1_+_239266921 | 0.03 |
ENSRNOT00000044486
|
Abhd17b
|
abhydrolase domain containing 17B |
| chr13_+_103396314 | 0.03 |
ENSRNOT00000049499
|
Slc30a10
|
solute carrier family 30, member 10 |
| chr5_+_63192298 | 0.02 |
ENSRNOT00000008329
|
Sec61b
|
Sec61 translocon beta subunit |
| chr10_-_56530842 | 0.02 |
ENSRNOT00000077451
|
AABR07029863.3
|
|
| chr7_-_98098268 | 0.02 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chr2_+_40000313 | 0.02 |
ENSRNOT00000014270
|
Depdc1b
|
DEP domain containing 1B |
| chr10_-_57638485 | 0.02 |
ENSRNOT00000009468
|
Dhx33
|
DEAH-box helicase 33 |
| chr1_+_266952561 | 0.02 |
ENSRNOT00000076452
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr18_+_65388685 | 0.02 |
ENSRNOT00000080844
|
Tcf4
|
transcription factor 4 |
| chr3_+_152909189 | 0.02 |
ENSRNOT00000066341
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr4_+_7258176 | 0.02 |
ENSRNOT00000061992
|
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr6_-_91676344 | 0.02 |
ENSRNOT00000083872
|
Nemf
|
nuclear export mediator factor |
| chr10_+_105156822 | 0.02 |
ENSRNOT00000012523
|
Galr2
|
galanin receptor 2 |
| chr15_+_45712821 | 0.02 |
ENSRNOT00000083381
ENSRNOT00000045833 |
Fam124a
|
family with sequence similarity 124 member A |
| chr1_-_220746224 | 0.02 |
ENSRNOT00000027734
|
Banf1
|
barrier to autointegration factor 1 |
| chr7_+_78005232 | 0.02 |
ENSRNOT00000005803
|
Dcaf13
|
DDB1 and CUL4 associated factor 13 |
| chr11_+_14005732 | 0.02 |
ENSRNOT00000047240
|
Rbm11
|
RNA binding motif protein 11 |
| chr10_+_47765432 | 0.02 |
ENSRNOT00000078231
|
LOC102553715
|
microfibril-associated glycoprotein 4-like |
| chr7_-_126465723 | 0.02 |
ENSRNOT00000021196
|
Wnt7b
|
wingless-type MMTV integration site family, member 7B |
| chr3_-_153468794 | 0.02 |
ENSRNOT00000082145
|
Ghrh
|
growth hormone releasing hormone |
| chr4_+_171748273 | 0.02 |
ENSRNOT00000009998
|
Dera
|
deoxyribose-phosphate aldolase |
| chr1_-_247978540 | 0.02 |
ENSRNOT00000039428
|
RGD1311595
|
similar to KIAA2026 protein |
| chr5_-_144274981 | 0.02 |
ENSRNOT00000065871
|
Map7d1
|
MAP7 domain containing 1 |
| chr3_-_94767239 | 0.02 |
ENSRNOT00000056808
|
Qser1
|
glutamine and serine rich 1 |
| chr2_+_242882306 | 0.02 |
ENSRNOT00000013661
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr13_-_83842051 | 0.02 |
ENSRNOT00000004376
|
Mpzl1
|
myelin protein zero-like 1 |
| chr10_-_57671080 | 0.02 |
ENSRNOT00000082511
|
LOC691995
|
hypothetical protein LOC691995 |
| chr6_+_135866739 | 0.02 |
ENSRNOT00000013460
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr6_+_132246602 | 0.02 |
ENSRNOT00000009896
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr15_+_19603288 | 0.02 |
ENSRNOT00000035491
|
LOC102552640
|
REST corepressor 2-like |
| chr1_-_84812486 | 0.02 |
ENSRNOT00000078369
|
AABR07002779.1
|
|
| chr6_+_69971227 | 0.02 |
ENSRNOT00000075349
|
Foxg1
|
forkhead box G1 |
| chr7_+_138707426 | 0.02 |
ENSRNOT00000037874
|
Pced1b
|
PC-esterase domain containing 1B |
| chr13_+_48790767 | 0.02 |
ENSRNOT00000087504
|
Elk4
|
ELK4, ETS transcription factor |
| chr16_+_73564243 | 0.02 |
ENSRNOT00000030960
|
Golga7
|
golgin A7 |
| chr10_+_49000973 | 0.02 |
ENSRNOT00000057880
|
LOC102553715
|
microfibril-associated glycoprotein 4-like |
| chr12_+_16340922 | 0.02 |
ENSRNOT00000001696
|
Snx8
|
sorting nexin 8 |
| chr10_+_47182433 | 0.02 |
ENSRNOT00000008679
|
LOC108348082
|
dual specificity mitogen-activated protein kinase kinase 3-like |
| chr11_+_61661724 | 0.02 |
ENSRNOT00000079423
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr3_-_153250641 | 0.02 |
ENSRNOT00000008961
|
Samhd1
|
SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 |
| chr16_+_71629525 | 0.02 |
ENSRNOT00000035347
ENSRNOT00000088462 |
Tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr13_-_88536728 | 0.02 |
ENSRNOT00000003950
|
Uhmk1
|
U2AF homology motif kinase 1 |
| chr12_+_22835019 | 0.02 |
ENSRNOT00000059530
|
Col26a1
|
collagen type XXVI alpha 1 chain |
| chr1_-_219544328 | 0.02 |
ENSRNOT00000025847
|
Grk2
|
G protein-coupled receptor kinase 2 |
| chr9_-_43022998 | 0.02 |
ENSRNOT00000063781
ENSRNOT00000089843 |
Lman2l
|
lectin, mannose-binding 2-like |
| chr4_+_10631073 | 0.02 |
ENSRNOT00000081939
ENSRNOT00000017907 |
Ptpn12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr11_+_34598492 | 0.02 |
ENSRNOT00000065600
|
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr8_+_56179816 | 0.02 |
ENSRNOT00000059078
|
Arhgap20
|
Rho GTPase activating protein 20 |
| chr8_-_104912959 | 0.02 |
ENSRNOT00000017363
|
Spsb4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr4_+_6931495 | 0.02 |
ENSRNOT00000079770
|
Wdr86
|
WD repeat domain 86 |
| chr1_-_188895223 | 0.02 |
ENSRNOT00000032796
|
Gpr139
|
G protein-coupled receptor 139 |
| chr18_+_70733872 | 0.02 |
ENSRNOT00000067018
|
Acaa2
|
acetyl-CoA acyltransferase 2 |
| chr1_+_140762758 | 0.02 |
ENSRNOT00000047848
|
Acan
|
aggrecan |
| chr2_-_149563905 | 0.02 |
ENSRNOT00000086186
|
Igsf10
|
immunoglobulin superfamily, member 10 |
| chr3_+_63536166 | 0.02 |
ENSRNOT00000016132
|
Plekha3
|
pleckstrin homology domain containing A3 |
| chr9_-_99299715 | 0.02 |
ENSRNOT00000027622
|
Hdac4
|
histone deacetylase 4 |
| chr6_+_127282466 | 0.02 |
ENSRNOT00000012124
|
Otub2
|
OTU deubiquitinase, ubiquitin aldehyde binding 2 |
| chr3_+_159569363 | 0.02 |
ENSRNOT00000064159
|
Tox2
|
TOX high mobility group box family member 2 |
| chr3_+_62648447 | 0.02 |
ENSRNOT00000002111
|
Agps
|
alkylglycerone phosphate synthase |
| chr3_-_7422738 | 0.02 |
ENSRNOT00000088339
|
Gtf3c4
|
general transcription factor IIIC subunit 4 |
| chr11_-_38351184 | 0.02 |
ENSRNOT00000046318
|
Prdm15
|
PR/SET domain 15 |
| chr16_+_86631064 | 0.02 |
ENSRNOT00000019675
|
Efnb2
|
ephrin B2 |
| chr9_-_46475040 | 0.02 |
ENSRNOT00000078196
|
Rfx8
|
RFX family member 8, lacking RFX DNA binding domain |
| chr12_+_7454884 | 0.02 |
ENSRNOT00000077328
|
LOC100910196
|
katanin p60 ATPase-containing subunit A-like 1-like |
| chr9_-_79898912 | 0.02 |
ENSRNOT00000022076
|
March4
|
membrane associated ring-CH-type finger 4 |
| chr6_+_122729874 | 0.02 |
ENSRNOT00000039509
ENSRNOT00000040440 |
Zc3h14
|
zinc finger CCCH type containing 14 |
| chr17_-_10575203 | 0.02 |
ENSRNOT00000073186
|
Arl10
|
ADP-ribosylation factor like GTPase 10 |
| chr17_-_22311392 | 0.02 |
ENSRNOT00000019389
|
Hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr17_+_70586394 | 0.02 |
ENSRNOT00000025550
|
Rbm17
|
RNA binding motif protein 17 |
| chr8_+_128133431 | 0.02 |
ENSRNOT00000020174
|
Exog
|
exo/endonuclease G |
| chr4_-_177331874 | 0.02 |
ENSRNOT00000065387
ENSRNOT00000091099 |
C2cd5
|
C2 calcium-dependent domain containing 5 |
| chr10_+_57185347 | 0.02 |
ENSRNOT00000040488
ENSRNOT00000086374 |
Mink1
|
misshapen-like kinase 1 |
| chr7_+_12006710 | 0.02 |
ENSRNOT00000045421
|
Klf16
|
Kruppel-like factor 16 |
| chr1_-_219853329 | 0.02 |
ENSRNOT00000026169
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr19_+_55929609 | 0.02 |
ENSRNOT00000068231
ENSRNOT00000077701 |
Cpne7
|
copine 7 |
| chr19_-_10826895 | 0.02 |
ENSRNOT00000090217
|
Rspry1
|
ring finger and SPRY domain containing 1 |
| chr10_+_10889488 | 0.02 |
ENSRNOT00000071746
|
Ubald1
|
UBA-like domain containing 1 |
| chr13_-_109552589 | 0.02 |
ENSRNOT00000005095
|
Vash2
|
vasohibin 2 |
| chr18_-_36285444 | 0.02 |
ENSRNOT00000087852
|
Prelid2
|
PRELI domain containing 2 |
| chr1_-_52992213 | 0.02 |
ENSRNOT00000033528
|
Prr18
|
proline rich 18 |
| chr4_-_16130848 | 0.02 |
ENSRNOT00000042914
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr10_+_10530365 | 0.02 |
ENSRNOT00000003843
|
Eef2kmt
|
eukaryotic elongation factor 2 lysine methyltransferase |
| chr9_+_94362299 | 0.02 |
ENSRNOT00000021168
|
Efhd1
|
EF-hand domain family, member D1 |
| chr10_+_13996645 | 0.02 |
ENSRNOT00000016490
|
Nthl1
|
nth-like DNA glycosylase 1 |
| chr8_-_59239954 | 0.02 |
ENSRNOT00000016104
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr1_+_224957517 | 0.02 |
ENSRNOT00000026033
|
Nxf1
|
nuclear RNA export factor 1 |
| chr6_+_42568220 | 0.02 |
ENSRNOT00000074829
|
Pdia6
|
protein disulfide isomerase family A, member 6 |
| chr1_-_21586910 | 0.02 |
ENSRNOT00000032424
|
Med23
|
mediator complex subunit 23 |
| chr17_-_10004321 | 0.02 |
ENSRNOT00000042394
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr13_+_50873605 | 0.02 |
ENSRNOT00000004382
|
Fmod
|
fibromodulin |
| chr10_+_57462447 | 0.02 |
ENSRNOT00000041443
|
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr4_+_175431904 | 0.01 |
ENSRNOT00000032843
|
Pde3a
|
phosphodiesterase 3A |
| chr1_+_100447998 | 0.01 |
ENSRNOT00000026259
|
Lrrc4b
|
leucine rich repeat containing 4B |
| chr11_-_84090259 | 0.01 |
ENSRNOT00000002321
|
Eif2b5
|
eukaryotic translation initiation factor 2B subunit 5 epsilon |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:0089718 | amino acid import across plasma membrane(GO:0089718) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.1 | GO:0097187 | odontoblast differentiation(GO:0071895) dentinogenesis(GO:0097187) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.0 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.1 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.0 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.0 | GO:0072054 | renal outer medulla development(GO:0072054) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.0 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.0 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |