Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
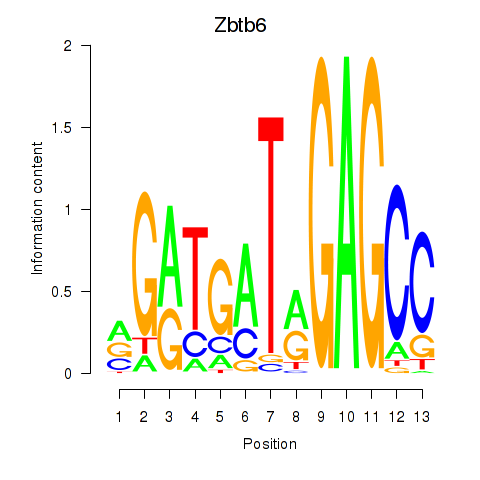
Results for Zbtb6
Z-value: 0.30
Transcription factors associated with Zbtb6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb6
|
ENSRNOG00000009340 | zinc finger and BTB domain containing 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb6 | rn6_v1_chr3_-_21684132_21684132 | 0.82 | 9.2e-02 | Click! |
Activity profile of Zbtb6 motif
Sorted Z-values of Zbtb6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_88215834 | 0.15 |
ENSRNOT00000034098
|
Gpr158
|
G protein-coupled receptor 158 |
| chr1_-_219412816 | 0.11 |
ENSRNOT00000083204
ENSRNOT00000029580 |
Rps6kb2
|
ribosomal protein S6 kinase B2 |
| chr2_+_198231291 | 0.10 |
ENSRNOT00000078288
ENSRNOT00000046739 |
Otud7b
|
OTU deubiquitinase 7B |
| chr7_+_23403891 | 0.10 |
ENSRNOT00000037918
|
Syn3
|
synapsin III |
| chr2_+_189714754 | 0.09 |
ENSRNOT00000086285
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr16_-_18937562 | 0.09 |
ENSRNOT00000080387
|
Nwd1
|
NACHT and WD repeat domain containing 1 |
| chr9_+_16003058 | 0.09 |
ENSRNOT00000081621
ENSRNOT00000021158 |
Ubr2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr3_-_125213607 | 0.09 |
ENSRNOT00000078070
|
Gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr17_-_1610745 | 0.09 |
ENSRNOT00000025850
|
Hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr10_+_94988362 | 0.09 |
ENSRNOT00000066525
|
Cep95
|
centrosomal protein 95 |
| chr2_-_165739874 | 0.08 |
ENSRNOT00000014384
|
Kpna4
|
karyopherin subunit alpha 4 |
| chrX_-_71313513 | 0.08 |
ENSRNOT00000004972
ENSRNOT00000076008 ENSRNOT00000076042 |
Zmym3
|
zinc finger MYM-type containing 3 |
| chr2_-_198002625 | 0.08 |
ENSRNOT00000091888
|
BC028528
|
cDNA sequence BC028528 |
| chr3_-_94182714 | 0.08 |
ENSRNOT00000015073
|
LOC100362814
|
hypothetical protein LOC100362814 |
| chrX_-_54303729 | 0.08 |
ENSRNOT00000087919
ENSRNOT00000064340 ENSRNOT00000051249 ENSRNOT00000087547 |
Gk
|
glycerol kinase |
| chr4_+_123801174 | 0.08 |
ENSRNOT00000029055
|
RGD1560289
|
similar to chromosome 3 open reading frame 20 |
| chr19_-_58735173 | 0.07 |
ENSRNOT00000030077
|
Pcnx2
|
pecanex homolog 2 (Drosophila) |
| chr1_+_214927172 | 0.07 |
ENSRNOT00000027134
|
Brsk2
|
BR serine/threonine kinase 2 |
| chr2_+_41911131 | 0.07 |
ENSRNOT00000016768
|
Plk2
|
polo-like kinase 2 |
| chr7_+_58419197 | 0.07 |
ENSRNOT00000085829
|
Zfc3h1
|
zinc finger, C3H1-type containing |
| chr1_-_241046249 | 0.07 |
ENSRNOT00000067796
ENSRNOT00000081866 |
Smc5
|
structural maintenance of chromosomes 5 |
| chr11_+_83884048 | 0.06 |
ENSRNOT00000002376
|
LOC108348101
|
chloride channel protein 2 |
| chr8_+_122003916 | 0.06 |
ENSRNOT00000091980
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chrX_+_22313028 | 0.06 |
ENSRNOT00000082725
|
Kdm5c
|
lysine demethylase 5C |
| chr10_+_34185898 | 0.06 |
ENSRNOT00000003339
|
Trim7
|
tripartite motif-containing 7 |
| chr7_+_78092037 | 0.06 |
ENSRNOT00000050753
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr9_-_81868086 | 0.05 |
ENSRNOT00000067080
|
Zfp142
|
zinc finger protein 142 |
| chr4_-_115157263 | 0.05 |
ENSRNOT00000015296
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr7_+_145117951 | 0.05 |
ENSRNOT00000055272
|
Pde1b
|
phosphodiesterase 1B |
| chr20_+_9791171 | 0.05 |
ENSRNOT00000078031
|
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chr17_-_45154355 | 0.05 |
ENSRNOT00000084976
|
Zkscan4
|
zinc finger with KRAB and SCAN domains 4 |
| chr8_+_122197027 | 0.05 |
ENSRNOT00000013050
|
Ubp1
|
upstream binding protein 1 (LBP-1a) |
| chr11_+_83048636 | 0.05 |
ENSRNOT00000002408
|
RGD1562339
|
RGD1562339 |
| chr19_-_53688597 | 0.05 |
ENSRNOT00000074448
|
Zcchc14
|
zinc finger CCHC-type containing 14 |
| chrX_-_22440187 | 0.05 |
ENSRNOT00000090731
|
Gpr173
|
|
| chr4_-_55713620 | 0.05 |
ENSRNOT00000010246
|
Gcc1
|
GRIP and coiled-coil domain containing 1 |
| chr13_+_51384562 | 0.05 |
ENSRNOT00000077928
|
Kdm5b
|
lysine demethylase 5B |
| chr18_-_48720472 | 0.05 |
ENSRNOT00000023236
|
Cep120
|
centrosomal protein 120 |
| chr16_+_70644474 | 0.05 |
ENSRNOT00000045955
|
LOC100359503
|
ribosomal protein S28-like |
| chr20_-_53777159 | 0.05 |
ENSRNOT00000073994
|
AABR07045621.1
|
|
| chr1_-_163129641 | 0.05 |
ENSRNOT00000083055
|
Capn5
|
calpain 5 |
| chr11_+_82862695 | 0.05 |
ENSRNOT00000071707
ENSRNOT00000073435 |
Clcn2
|
chloride voltage-gated channel 2 |
| chr15_-_82482009 | 0.05 |
ENSRNOT00000011926
|
Dach1
|
dachshund family transcription factor 1 |
| chr2_-_177924970 | 0.05 |
ENSRNOT00000029340
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr7_+_116980076 | 0.04 |
ENSRNOT00000012321
|
Zfp623
|
zinc finger protein 623 |
| chr14_-_114484127 | 0.04 |
ENSRNOT00000056706
|
Eml6
|
echinoderm microtubule associated protein like 6 |
| chr18_+_17403407 | 0.04 |
ENSRNOT00000045150
|
RGD1562608
|
similar to KIAA1328 protein |
| chr4_+_147832136 | 0.04 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr20_-_4934871 | 0.04 |
ENSRNOT00000071840
|
RT1-CE3
|
RT1 class I, locus CE3 |
| chr16_+_7435634 | 0.04 |
ENSRNOT00000026323
|
Capn7
|
calpain 7 |
| chr6_-_24563246 | 0.04 |
ENSRNOT00000074294
|
LOC685881
|
hypothetical protein LOC685881 |
| chr11_+_83883879 | 0.04 |
ENSRNOT00000050927
|
LOC108348101
|
chloride channel protein 2 |
| chr10_-_63491713 | 0.04 |
ENSRNOT00000063941
|
Tusc5
|
tumor suppressor candidate 5 |
| chr17_+_45078556 | 0.04 |
ENSRNOT00000088280
|
Zfp192
|
zinc finger protein 192 |
| chr20_+_2501252 | 0.04 |
ENSRNOT00000079307
ENSRNOT00000084559 |
Trim39
|
tripartite motif-containing 39 |
| chr18_+_30435119 | 0.04 |
ENSRNOT00000027190
|
Pcdhb8
|
protocadherin beta 8 |
| chr8_+_52753011 | 0.04 |
ENSRNOT00000047264
|
Nxpe1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr10_+_68588789 | 0.04 |
ENSRNOT00000049614
|
AABR07030122.1
|
|
| chr17_-_48562838 | 0.04 |
ENSRNOT00000017102
ENSRNOT00000084702 |
Amph
|
amphiphysin |
| chr14_+_3204806 | 0.04 |
ENSRNOT00000061583
|
Glmn
|
glomulin, FKBP associated protein |
| chr5_+_128215711 | 0.04 |
ENSRNOT00000011670
|
Cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr7_+_137155481 | 0.04 |
ENSRNOT00000089895
|
Ano6
|
anoctamin 6 |
| chr14_+_83510278 | 0.04 |
ENSRNOT00000081161
|
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr10_-_51669297 | 0.04 |
ENSRNOT00000071595
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr10_+_63677396 | 0.04 |
ENSRNOT00000005100
|
Slc43a2
|
solute carrier family 43 member 2 |
| chrX_+_40258493 | 0.04 |
ENSRNOT00000033010
|
Mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr7_-_2961873 | 0.04 |
ENSRNOT00000067441
|
Rps15-ps2
|
ribosomal protein S15, pseudogene 2 |
| chr10_-_86554478 | 0.03 |
ENSRNOT00000049621
|
Ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr4_+_170347410 | 0.03 |
ENSRNOT00000040508
|
LOC500350
|
LRRGT00139 |
| chr9_+_100885051 | 0.03 |
ENSRNOT00000082987
|
AC109427.1
|
|
| chr7_-_140172448 | 0.03 |
ENSRNOT00000077672
|
LOC103692976
|
cyclin-T1-like |
| chr10_+_29289203 | 0.03 |
ENSRNOT00000067013
|
Pwwp2a
|
PWWP domain containing 2A |
| chr12_+_11433813 | 0.03 |
ENSRNOT00000031901
ENSRNOT00000084577 |
Smurf1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr13_-_52256196 | 0.03 |
ENSRNOT00000011105
|
Ipo9
|
importin 9 |
| chr10_+_63659085 | 0.03 |
ENSRNOT00000005039
|
Rilp
|
Rab interacting lysosomal protein |
| chr5_-_135994848 | 0.03 |
ENSRNOT00000067675
|
Btbd19
|
BTB domain containing 19 |
| chr3_+_152402520 | 0.03 |
ENSRNOT00000027086
|
Cnbd2
|
cyclic nucleotide binding domain containing 2 |
| chr10_+_84966989 | 0.03 |
ENSRNOT00000013580
|
Scrn2
|
secernin 2 |
| chr1_+_255040426 | 0.03 |
ENSRNOT00000092151
|
Pcgf5
|
polycomb group ring finger 5 |
| chrX_+_2435305 | 0.03 |
ENSRNOT00000005623
|
Slc9a7
|
solute carrier family 9 member A7 |
| chr2_+_3400977 | 0.03 |
ENSRNOT00000093593
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr18_-_56331991 | 0.03 |
ENSRNOT00000085841
ENSRNOT00000091220 |
Slc6a7
|
solute carrier family 6 member 7 |
| chr12_-_11783232 | 0.03 |
ENSRNOT00000075888
|
Zfp498
|
zinc finger protein 498 |
| chrX_-_70428364 | 0.03 |
ENSRNOT00000045907
|
P2ry4
|
pyrimidinergic receptor P2Y4 |
| chr7_-_12762341 | 0.03 |
ENSRNOT00000064886
|
Abca7
|
ATP binding cassette subfamily A member 7 |
| chr8_+_117620317 | 0.03 |
ENSRNOT00000084220
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr4_+_13405136 | 0.03 |
ENSRNOT00000091004
|
Gnai1
|
G protein subunit alpha i1 |
| chr18_-_31071371 | 0.03 |
ENSRNOT00000060432
|
Diaph1
|
diaphanous-related formin 1 |
| chr3_-_29996865 | 0.03 |
ENSRNOT00000080382
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr10_-_90410569 | 0.03 |
ENSRNOT00000036112
|
Itga2b
|
integrin subunit alpha 2b |
| chr19_+_37568113 | 0.02 |
ENSRNOT00000023710
|
Fam65a
|
family with sequence similarity 65, member A |
| chr12_-_23925741 | 0.02 |
ENSRNOT00000001957
|
Srrm3
|
serine/arginine repetitive matrix 3 |
| chr7_+_2605719 | 0.02 |
ENSRNOT00000018737
|
Gls2
|
glutaminase 2 |
| chr5_-_6186329 | 0.02 |
ENSRNOT00000012610
|
Sulf1
|
sulfatase 1 |
| chr1_-_82088275 | 0.02 |
ENSRNOT00000072616
|
Dedd2
|
death effector domain containing 2 |
| chr4_+_10823281 | 0.02 |
ENSRNOT00000066739
|
Phtf2
|
putative homeodomain transcription factor 2 |
| chr1_-_90151405 | 0.02 |
ENSRNOT00000028688
|
RGD1308428
|
similar to RIKEN cDNA 4931406P16 |
| chr3_-_15278645 | 0.02 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr6_-_144123596 | 0.02 |
ENSRNOT00000006144
ENSRNOT00000087845 |
Wdr60
|
WD repeat domain 60 |
| chr20_-_4935372 | 0.02 |
ENSRNOT00000050099
ENSRNOT00000047779 |
RT1-CE3
RT1-CE4
|
RT1 class I, locus CE3 RT1 class I, locus CE4 |
| chr8_+_64610952 | 0.02 |
ENSRNOT00000015963
|
Myo9a
|
myosin IXA |
| chr1_-_214265668 | 0.02 |
ENSRNOT00000024094
|
Sct
|
secretin |
| chr11_+_86903122 | 0.02 |
ENSRNOT00000048063
|
Zdhhc8
|
zinc finger, DHHC-type containing 8 |
| chr11_+_64761146 | 0.02 |
ENSRNOT00000051428
|
Poglut1
|
protein O-glucosyltransferase 1 |
| chr8_-_111101416 | 0.02 |
ENSRNOT00000084413
|
Cep63
|
centrosomal protein 63 |
| chr19_-_10596851 | 0.02 |
ENSRNOT00000021716
|
Coq9
|
coenzyme Q9 |
| chr7_+_116653664 | 0.02 |
ENSRNOT00000076606
|
Zfp41
|
zinc finger protein 41 |
| chr2_-_157759819 | 0.02 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chr10_+_84986328 | 0.02 |
ENSRNOT00000085830
ENSRNOT00000067665 |
Osbpl7
|
oxysterol binding protein-like 7 |
| chr2_+_220432037 | 0.02 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr10_-_84847857 | 0.02 |
ENSRNOT00000071495
|
LOC102552549
|
migration and invasion enhancer 1-like |
| chr8_+_48382121 | 0.02 |
ENSRNOT00000008685
|
Thy1
|
Thy-1 cell surface antigen |
| chr10_-_49196177 | 0.02 |
ENSRNOT00000084418
|
Zfp286a
|
zinc finger protein 286A |
| chr8_+_115069095 | 0.02 |
ENSRNOT00000014770
|
Dusp7
|
dual specificity phosphatase 7 |
| chr8_-_63350269 | 0.01 |
ENSRNOT00000042681
|
Cd276
|
Cd276 molecule |
| chr1_-_31122093 | 0.01 |
ENSRNOT00000016712
|
NEWGENE_1307525
|
SOGA family member 3 |
| chr14_+_25589762 | 0.01 |
ENSRNOT00000043938
ENSRNOT00000067439 ENSRNOT00000002793 |
Epha5
|
EPH receptor A5 |
| chr14_+_83510640 | 0.01 |
ENSRNOT00000089928
ENSRNOT00000082242 ENSRNOT00000025378 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr19_-_19896421 | 0.01 |
ENSRNOT00000020793
|
Heatr3
|
HEAT repeat containing 3 |
| chr12_+_37603986 | 0.01 |
ENSRNOT00000088006
|
Sbno1
|
strawberry notch homolog 1 |
| chr11_+_87366621 | 0.01 |
ENSRNOT00000040978
|
Aifm3
|
apoptosis inducing factor, mitochondria associated 3 |
| chr14_-_83741969 | 0.01 |
ENSRNOT00000026293
|
Inpp5j
|
inositol polyphosphate-5-phosphatase J |
| chr6_+_134804141 | 0.01 |
ENSRNOT00000086143
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_+_255479261 | 0.01 |
ENSRNOT00000079808
|
Tnks2
|
tankyrase 2 |
| chr1_-_222224910 | 0.01 |
ENSRNOT00000028720
|
Plcb3
|
phospholipase C beta 3 |
| chr3_+_92403582 | 0.01 |
ENSRNOT00000064282
|
Pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr8_-_94368834 | 0.01 |
ENSRNOT00000078977
|
Me1
|
malic enzyme 1 |
| chr1_-_131454689 | 0.01 |
ENSRNOT00000014152
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr15_-_28733513 | 0.01 |
ENSRNOT00000078180
|
Sall2
|
spalt-like transcription factor 2 |
| chr10_-_27366665 | 0.01 |
ENSRNOT00000004725
|
Gabra1
|
gamma-aminobutyric acid type A receptor alpha1 subunit |
| chr15_+_62520953 | 0.01 |
ENSRNOT00000040851
|
LOC306079
|
similar to RIKEN cDNA 3100001N19 |
| chr2_-_185005572 | 0.01 |
ENSRNOT00000093291
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr5_+_123905166 | 0.01 |
ENSRNOT00000082021
|
Dab1
|
DAB1, reelin adaptor protein |
| chr8_-_94120433 | 0.01 |
ENSRNOT00000080195
ENSRNOT00000014600 |
Ube3d
|
ubiquitin protein ligase E3D |
| chr10_+_78168715 | 0.01 |
ENSRNOT00000072294
|
Cox11
|
COX11 cytochrome c oxidase copper chaperone |
| chr5_-_62621737 | 0.01 |
ENSRNOT00000011573
|
Gabbr2
|
gamma-aminobutyric acid type B receptor subunit 2 |
| chr13_+_71086745 | 0.01 |
ENSRNOT00000083366
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr14_-_3204588 | 0.01 |
ENSRNOT00000002815
|
Rpap2
|
RNA polymerase II associated protein 2 |
| chr5_-_50142724 | 0.01 |
ENSRNOT00000081150
|
Slc35a1
|
solute carrier family 35 member A1 |
| chr16_-_19308842 | 0.01 |
ENSRNOT00000019556
|
Fam32a
|
family with sequence similarity 32, member A |
| chr11_+_60613882 | 0.01 |
ENSRNOT00000002853
|
Slc35a5
|
solute carrier family 35, member A5 |
| chrX_+_114929029 | 0.01 |
ENSRNOT00000006459
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr10_-_70646495 | 0.01 |
ENSRNOT00000071218
|
Gas2l2
|
growth arrest-specific 2 like 2 |
| chr7_-_23948202 | 0.01 |
ENSRNOT00000044252
|
Ascl4
|
achaete-scute family bHLH transcription factor 4 |
| chr2_-_188689392 | 0.01 |
ENSRNOT00000027986
|
Dcst1
|
DC-STAMP domain containing 1 |
| chr5_+_172275734 | 0.01 |
ENSRNOT00000064266
|
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr20_-_4530126 | 0.01 |
ENSRNOT00000000481
|
Skiv2l
|
Ski2 like RNA helicase |
| chr12_-_24578761 | 0.01 |
ENSRNOT00000076209
|
Tbl2
|
transducin (beta)-like 2 |
| chr8_+_70447040 | 0.01 |
ENSRNOT00000016798
|
Ints14
|
integrator complex subunit 14 |
| chr10_+_35279236 | 0.01 |
ENSRNOT00000003770
|
Gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr10_-_82197520 | 0.01 |
ENSRNOT00000092024
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr10_-_103816287 | 0.00 |
ENSRNOT00000004477
|
Grin2c
|
glutamate ionotropic receptor NMDA type subunit 2C |
| chr5_+_156615886 | 0.00 |
ENSRNOT00000076439
|
Sh2d5
|
SH2 domain containing 5 |
| chr10_-_78168609 | 0.00 |
ENSRNOT00000041119
|
Stxbp4
|
syntaxin binding protein 4 |
| chr1_-_42971208 | 0.00 |
ENSRNOT00000088535
|
Gm5414
|
predicted gene 5414 |
| chr12_-_13998172 | 0.00 |
ENSRNOT00000001476
|
Wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr3_-_51890749 | 0.00 |
ENSRNOT00000088405
|
AABR07052405.1
|
|
| chrX_-_10413984 | 0.00 |
ENSRNOT00000039551
ENSRNOT00000091448 |
Ddx3x
|
DEAD-box helicase 3, X-linked |
| chr6_-_33738825 | 0.00 |
ENSRNOT00000039492
|
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr16_-_71319449 | 0.00 |
ENSRNOT00000029284
|
Fgfr1
|
Fibroblast growth factor receptor 1 |
| chr18_-_6474990 | 0.00 |
ENSRNOT00000061504
|
Kctd1
|
potassium channel tetramerization domain containing 1 |
| chr16_-_47537476 | 0.00 |
ENSRNOT00000050279
|
Cldn24
|
claudin 24 |
| chr1_-_259106948 | 0.00 |
ENSRNOT00000074429
|
LOC100912537
|
tectonic-3-like |
| chr16_-_49522341 | 0.00 |
ENSRNOT00000081642
|
LOC100911140
|
coiled-coil domain-containing protein 110-like |
| chr4_+_157612536 | 0.00 |
ENSRNOT00000055970
|
Chd4
|
chromodomain helicase DNA binding protein 4 |
| chr7_-_134722215 | 0.00 |
ENSRNOT00000036750
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr13_+_51022681 | 0.00 |
ENSRNOT00000078599
|
Chi3l1
|
chitinase 3 like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0017143 | insecticide metabolic process(GO:0017143) |
| 0.0 | 0.1 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 0.1 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.0 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.0 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.0 | GO:0035938 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.0 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.0 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |