Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
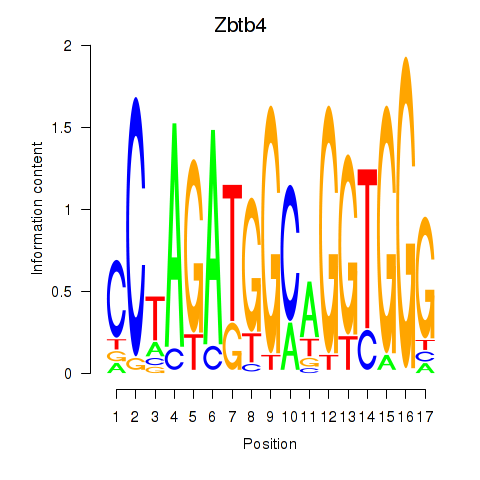
Results for Zbtb4
Z-value: 0.48
Transcription factors associated with Zbtb4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb4
|
ENSRNOG00000014689 | zinc finger and BTB domain containing 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb4 | rn6_v1_chr10_+_56381813_56381813 | -0.67 | 2.2e-01 | Click! |
Activity profile of Zbtb4 motif
Sorted Z-values of Zbtb4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_45815940 | 0.35 |
ENSRNOT00000073276
|
Gpr6
|
G protein-coupled receptor 6 |
| chrX_+_136460215 | 0.17 |
ENSRNOT00000093538
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr1_-_80783898 | 0.14 |
ENSRNOT00000045306
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr2_-_195888216 | 0.14 |
ENSRNOT00000056436
|
Tuft1
|
tuftelin 1 |
| chr4_+_114935970 | 0.13 |
ENSRNOT00000082336
|
Slc4a5
|
solute carrier family 4 member 5 |
| chr10_-_90049112 | 0.13 |
ENSRNOT00000028323
|
Pyy
|
peptide YY (mapped) |
| chr4_+_154309426 | 0.12 |
ENSRNOT00000019346
|
A2m
|
alpha-2-macroglobulin |
| chr7_-_130350570 | 0.12 |
ENSRNOT00000055805
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr8_-_55491152 | 0.12 |
ENSRNOT00000014965
|
LOC100359479
|
rCG58364-like |
| chr7_+_13062196 | 0.11 |
ENSRNOT00000000193
|
Plpp2
|
phospholipid phosphatase 2 |
| chr3_+_65815080 | 0.10 |
ENSRNOT00000006429
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr20_+_2004052 | 0.10 |
ENSRNOT00000001008
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr2_+_211930326 | 0.10 |
ENSRNOT00000027765
|
Slc25a24
|
solute carrier family 25 member 24 |
| chr9_-_64096265 | 0.09 |
ENSRNOT00000013502
|
Tyw5
|
tRNA-yW synthesizing protein 5 |
| chr11_+_32211115 | 0.09 |
ENSRNOT00000087452
|
Mrps6
|
mitochondrial ribosomal protein S6 |
| chr2_+_211337271 | 0.08 |
ENSRNOT00000045155
|
Cox6b1
|
cytochrome c oxidase subunit 6B1 |
| chr7_-_115979298 | 0.08 |
ENSRNOT00000075883
|
RGD1308195
|
similar to secreted Ly6/uPAR related protein 2 |
| chr8_-_6235967 | 0.08 |
ENSRNOT00000068290
|
LOC654482
|
hypothetical protein LOC654482 |
| chr10_+_56445647 | 0.08 |
ENSRNOT00000056870
|
Tmem256
|
transmembrane protein 256 |
| chr8_+_65686648 | 0.08 |
ENSRNOT00000017233
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr13_-_102721218 | 0.08 |
ENSRNOT00000005459
|
Marc1
|
mitochondrial amidoxime reducing component 1 |
| chr16_+_69048730 | 0.08 |
ENSRNOT00000086082
ENSRNOT00000078128 |
Rab11fip1
|
RAB11 family interacting protein 1 |
| chr5_+_138154673 | 0.08 |
ENSRNOT00000064452
|
Slc2a1
|
solute carrier family 2 member 1 |
| chr6_+_95826546 | 0.08 |
ENSRNOT00000043587
|
AABR07064810.1
|
|
| chr1_-_91588609 | 0.07 |
ENSRNOT00000050931
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr2_+_84678948 | 0.07 |
ENSRNOT00000046325
|
Fam173b
|
family with sequence similarity 173, member B |
| chr8_-_111965889 | 0.07 |
ENSRNOT00000032376
|
Bfsp2
|
beaded filament structural protein 2 |
| chr1_+_240908483 | 0.07 |
ENSRNOT00000019367
|
Klf9
|
Kruppel-like factor 9 |
| chr3_-_9738752 | 0.07 |
ENSRNOT00000045993
|
Ptges
|
prostaglandin E synthase |
| chr14_+_89314176 | 0.07 |
ENSRNOT00000006765
|
Upp1
|
uridine phosphorylase 1 |
| chr15_-_52332097 | 0.07 |
ENSRNOT00000017723
|
Fgf17
|
fibroblast growth factor 17 |
| chr4_-_84768249 | 0.07 |
ENSRNOT00000013205
|
Fkbp14
|
FK506 binding protein 14 |
| chr5_+_159845774 | 0.07 |
ENSRNOT00000012328
|
Epha2
|
Eph receptor A2 |
| chr15_-_61564695 | 0.07 |
ENSRNOT00000068216
|
Rgcc
|
regulator of cell cycle |
| chr1_-_162385575 | 0.06 |
ENSRNOT00000016540
|
Thrsp
|
thyroid hormone responsive |
| chr3_-_15433252 | 0.06 |
ENSRNOT00000008817
|
Lhx6
|
LIM homeobox 6 |
| chr5_-_16706909 | 0.06 |
ENSRNOT00000011314
|
Rps20
|
ribosomal protein S20 |
| chr1_+_193335645 | 0.06 |
ENSRNOT00000019640
|
Lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr11_+_81358592 | 0.06 |
ENSRNOT00000002487
|
Rfc4
|
replication factor C subunit 4 |
| chr4_+_154423209 | 0.06 |
ENSRNOT00000075799
|
LOC100911545
|
alpha-2-macroglobulin-like |
| chr20_+_3556975 | 0.06 |
ENSRNOT00000089417
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_-_83977236 | 0.05 |
ENSRNOT00000028434
|
Egln2
|
egl-9 family hypoxia-inducible factor 2 |
| chr7_+_10962330 | 0.05 |
ENSRNOT00000008360
|
Tle6
|
transducin-like enhancer of split 6 |
| chr8_-_53362006 | 0.05 |
ENSRNOT00000077145
|
LOC100360390
|
claudin 25-like |
| chr19_+_33928356 | 0.05 |
ENSRNOT00000058162
|
Ednra
|
endothelin receptor type A |
| chr5_-_136721379 | 0.05 |
ENSRNOT00000026704
|
Atp6v0b
|
ATPase H+ transporting V0 subunit B |
| chr5_+_126812187 | 0.05 |
ENSRNOT00000057372
|
Ldlrad1
|
low density lipoprotein receptor class A domain containing 1 |
| chr5_-_50193571 | 0.05 |
ENSRNOT00000051243
|
Cfap206
|
cilia and flagella associated protein 206 |
| chr9_+_101284902 | 0.05 |
ENSRNOT00000065533
|
NEWGENE_1308624
|
sialidase 4 |
| chr18_-_55859333 | 0.05 |
ENSRNOT00000025989
|
Myoz3
|
myozenin 3 |
| chr7_+_122700854 | 0.05 |
ENSRNOT00000086603
|
Rbx1
|
ring-box 1 |
| chr5_+_48011864 | 0.05 |
ENSRNOT00000067610
|
Mdn1
|
midasin AAA ATPase 1 |
| chr9_+_10535340 | 0.05 |
ENSRNOT00000075408
|
Znrf4
|
zinc and ring finger 4 |
| chr2_+_187218851 | 0.05 |
ENSRNOT00000017798
|
Sh2d2a
|
SH2 domain containing 2A |
| chr1_+_7101715 | 0.05 |
ENSRNOT00000019994
|
Sf3b5
|
splicing factor 3b, subunit 5 |
| chrX_+_124516949 | 0.05 |
ENSRNOT00000051474
|
Rnf113a1
|
ring finger protein 113A1 |
| chr7_-_59514939 | 0.05 |
ENSRNOT00000085579
|
Kcnmb4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr15_+_52241801 | 0.05 |
ENSRNOT00000082639
|
Hr
|
HR, lysine demethylase and nuclear receptor corepressor |
| chr2_-_84678790 | 0.05 |
ENSRNOT00000015886
|
Cct5
|
chaperonin containing TCP1 subunit 5 |
| chr5_+_79179417 | 0.05 |
ENSRNOT00000010454
|
Orm1
|
orosomucoid 1 |
| chr10_-_45851984 | 0.05 |
ENSRNOT00000058308
|
Zfp496
|
zinc finger protein 496 |
| chr15_+_61879184 | 0.05 |
ENSRNOT00000042606
|
Sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr5_+_169364306 | 0.05 |
ENSRNOT00000014213
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr19_+_16417817 | 0.04 |
ENSRNOT00000015272
|
Irx5
|
iroquois homeobox 5 |
| chr4_-_171176581 | 0.04 |
ENSRNOT00000030850
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr2_-_187622373 | 0.04 |
ENSRNOT00000026396
|
Rhbg
|
Rh family B glycoprotein |
| chrX_+_143097525 | 0.04 |
ENSRNOT00000004559
|
F9
|
coagulation factor IX |
| chr1_+_189870622 | 0.04 |
ENSRNOT00000075003
|
Tmem159
|
transmembrane protein 159 |
| chr9_+_80118029 | 0.04 |
ENSRNOT00000023068
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr5_+_152559577 | 0.04 |
ENSRNOT00000082245
|
Slc30a2
|
solute carrier family 30 member 2 |
| chr10_-_62032400 | 0.04 |
ENSRNOT00000086378
ENSRNOT00000004161 |
Dph1
|
diphthamide biosynthesis 1 |
| chr16_+_74292438 | 0.04 |
ENSRNOT00000026197
|
Vdac3
|
voltage-dependent anion channel 3 |
| chrX_+_123404518 | 0.04 |
ENSRNOT00000015085
|
Slc25a5
|
solute carrier family 25 member 5 |
| chr8_+_13796021 | 0.04 |
ENSRNOT00000013927
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr19_+_52374298 | 0.04 |
ENSRNOT00000092131
|
Atp2c2
|
ATPase secretory pathway Ca2+ transporting 2 |
| chr10_-_49196177 | 0.04 |
ENSRNOT00000084418
|
Zfp286a
|
zinc finger protein 286A |
| chr1_-_225631468 | 0.04 |
ENSRNOT00000072579
|
AABR07006232.1
|
|
| chr12_+_12227010 | 0.04 |
ENSRNOT00000060843
ENSRNOT00000092610 |
Baiap2l1
|
BAI1-associated protein 2-like 1 |
| chr3_+_46286712 | 0.04 |
ENSRNOT00000085563
|
March7
|
membrane associated ring-CH-type finger 7 |
| chr4_-_123494742 | 0.04 |
ENSRNOT00000073268
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr11_-_45510961 | 0.04 |
ENSRNOT00000002238
|
Tomm70
|
translocase of outer mitochondrial membrane 70 |
| chr1_-_101741441 | 0.04 |
ENSRNOT00000028570
|
Sult2b1
|
sulfotransferase family 2B member 1 |
| chr12_+_52452273 | 0.04 |
ENSRNOT00000056680
ENSRNOT00000088381 |
Pxmp2
|
peroxisomal membrane protein 2 |
| chrX_+_20351486 | 0.04 |
ENSRNOT00000093675
ENSRNOT00000047444 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr18_+_70263359 | 0.04 |
ENSRNOT00000019573
|
Cfap53
|
cilia and flagella associated protein 53 |
| chr17_-_9441526 | 0.04 |
ENSRNOT00000000146
|
Txndc15
|
thioredoxin domain containing 15 |
| chr4_+_7282948 | 0.03 |
ENSRNOT00000011052
|
Cdk5
|
cyclin-dependent kinase 5 |
| chr10_+_47282208 | 0.03 |
ENSRNOT00000057953
|
Kcnj12
|
potassium voltage-gated channel subfamily J member 12 |
| chr7_+_12179203 | 0.03 |
ENSRNOT00000049170
|
Mbd3
|
methyl-CpG binding domain protein 3 |
| chr4_-_115239723 | 0.03 |
ENSRNOT00000042699
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr9_-_121972055 | 0.03 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr1_-_263803150 | 0.03 |
ENSRNOT00000017840
|
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr1_-_100537377 | 0.03 |
ENSRNOT00000026599
|
Spib
|
Spi-B transcription factor |
| chr20_-_2040422 | 0.03 |
ENSRNOT00000052311
|
RT1-M5
|
RT1 class Ib, locus M5 |
| chr20_+_17750744 | 0.03 |
ENSRNOT00000000745
|
RGD1559903
|
similar to RIKEN cDNA 1700049L16 |
| chr1_-_214511529 | 0.03 |
ENSRNOT00000026390
|
Chid1
|
chitinase domain containing 1 |
| chr1_+_101704746 | 0.03 |
ENSRNOT00000028564
|
Fam83e
|
family with sequence similarity 83, member E |
| chr6_+_137210052 | 0.03 |
ENSRNOT00000031788
ENSRNOT00000065213 |
Siva1
|
SIVA1, apoptosis-inducing factor |
| chr16_+_56247659 | 0.03 |
ENSRNOT00000017452
|
Tusc3
|
tumor suppressor candidate 3 |
| chr1_-_53038229 | 0.03 |
ENSRNOT00000017282
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr1_-_81412251 | 0.03 |
ENSRNOT00000026946
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr10_-_38969501 | 0.03 |
ENSRNOT00000090691
ENSRNOT00000081309 ENSRNOT00000010029 |
Il4
|
interleukin 4 |
| chr4_-_80367606 | 0.03 |
ENSRNOT00000014331
|
LOC500124
|
similar to RIKEN cDNA 4921507P07 |
| chr5_-_131860637 | 0.03 |
ENSRNOT00000064569
ENSRNOT00000080242 |
Slc5a9
|
solute carrier family 5 member 9 |
| chr1_-_266142538 | 0.03 |
ENSRNOT00000026792
|
Actr1a
|
ARP1 actin-related protein 1 homolog A, centractin alpha |
| chr8_+_114897011 | 0.03 |
ENSRNOT00000074683
|
Twf2
|
twinfilin actin-binding protein 2 |
| chr13_+_78805347 | 0.03 |
ENSRNOT00000003748
|
Serpinc1
|
serpin family C member 1 |
| chr18_-_51651267 | 0.03 |
ENSRNOT00000020325
|
Aldh7a1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr8_-_126495347 | 0.03 |
ENSRNOT00000013467
|
Cmc1
|
C-x(9)-C motif containing 1 |
| chr9_-_65879521 | 0.03 |
ENSRNOT00000017517
|
Als2cr11
|
amyotrophic lateral sclerosis 2 chromosome region, candidate 11 |
| chr4_-_155631856 | 0.03 |
ENSRNOT00000088270
|
Slc2a3
|
solute carrier family 2 member 3 |
| chr10_+_13061170 | 0.03 |
ENSRNOT00000004936
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr19_-_869490 | 0.03 |
ENSRNOT00000080433
|
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chrX_+_114929029 | 0.03 |
ENSRNOT00000006459
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr18_+_29506128 | 0.03 |
ENSRNOT00000003878
|
Slc35a4
|
solute carrier family 35, member A4 |
| chr7_+_36826360 | 0.03 |
ENSRNOT00000029783
|
Ndufa13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| chr1_-_102970950 | 0.03 |
ENSRNOT00000018194
|
Tsg101
|
tumor susceptibility 101 |
| chr1_+_197813963 | 0.03 |
ENSRNOT00000045531
|
LOC100361060
|
ribosomal protein L36-like |
| chr16_+_56248331 | 0.03 |
ENSRNOT00000085300
|
Tusc3
|
tumor suppressor candidate 3 |
| chr1_+_224970807 | 0.03 |
ENSRNOT00000026141
|
Tmem223
|
transmembrane protein 223 |
| chr18_+_73645907 | 0.03 |
ENSRNOT00000023487
|
Loxhd1
|
lipoxygenase homology domains 1 |
| chr9_+_64095978 | 0.03 |
ENSRNOT00000021533
|
Maip1
|
matrix AAA peptidase interacting protein 1 |
| chr7_+_12247498 | 0.02 |
ENSRNOT00000022358
|
Pcsk4
|
proprotein convertase subtilisin/kexin type 4 |
| chr14_-_18849258 | 0.02 |
ENSRNOT00000033406
|
Pf4
|
platelet factor 4 |
| chr1_-_214074663 | 0.02 |
ENSRNOT00000026215
|
LOC100911881
|
chitinase domain-containing protein 1-like |
| chr1_-_100980340 | 0.02 |
ENSRNOT00000064272
|
Prmt1
|
protein arginine methyltransferase 1 |
| chr16_-_7007287 | 0.02 |
ENSRNOT00000041216
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr1_+_85496937 | 0.02 |
ENSRNOT00000076809
ENSRNOT00000087175 |
Selenov
|
selenoprotein V |
| chr7_-_119716238 | 0.02 |
ENSRNOT00000075678
|
Il2rb
|
interleukin 2 receptor subunit beta |
| chr7_-_138707221 | 0.02 |
ENSRNOT00000009199
|
Amigo2
|
adhesion molecule with Ig like domain 2 |
| chr1_-_170397191 | 0.02 |
ENSRNOT00000090181
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr5_+_19471664 | 0.02 |
ENSRNOT00000013563
|
Sdcbp
|
syndecan binding protein |
| chr7_-_123156558 | 0.02 |
ENSRNOT00000005943
|
Polr3h
|
RNA polymerase III subunit H |
| chr17_+_23986758 | 0.02 |
ENSRNOT00000024066
|
Sirt5
|
sirtuin 5 |
| chr10_+_49299125 | 0.02 |
ENSRNOT00000074853
|
Tvp23b
|
trans-golgi network vesicle protein 23 homolog B |
| chr7_+_138707426 | 0.02 |
ENSRNOT00000037874
|
Pced1b
|
PC-esterase domain containing 1B |
| chr1_-_198128857 | 0.02 |
ENSRNOT00000026496
|
Coro1a
|
coronin 1A |
| chr5_+_58151985 | 0.02 |
ENSRNOT00000020885
|
Il11ra1
|
interleukin 11 receptor subunit alpha 1 |
| chr19_+_61677542 | 0.02 |
ENSRNOT00000014785
|
Itgb1
|
integrin subunit beta 1 |
| chr3_+_55369214 | 0.02 |
ENSRNOT00000067161
|
Nostrin
|
nitric oxide synthase trafficking |
| chr10_-_105228547 | 0.02 |
ENSRNOT00000028875
|
Srp68
|
signal recognition particle 68 |
| chr4_+_61924013 | 0.02 |
ENSRNOT00000090717
|
Bpgm
|
bisphosphoglycerate mutase |
| chr8_+_59111271 | 0.02 |
ENSRNOT00000029413
|
Sh2d7
|
SH2 domain containing 7 |
| chr3_-_2456041 | 0.02 |
ENSRNOT00000062001
|
Rnf224
|
ring finger protein 224 |
| chr4_-_58875753 | 0.02 |
ENSRNOT00000016991
|
Podxl
|
podocalyxin-like |
| chr9_+_42871950 | 0.02 |
ENSRNOT00000089673
|
Arid5a
|
AT-rich interaction domain 5A |
| chr12_-_40680374 | 0.02 |
ENSRNOT00000048984
ENSRNOT00000080418 |
Naa25
|
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
| chr8_+_81766041 | 0.02 |
ENSRNOT00000032280
|
Onecut1
|
one cut homeobox 1 |
| chr19_-_19727081 | 0.02 |
ENSRNOT00000020700
|
Adcy7
|
adenylate cyclase 7 |
| chr17_+_77224112 | 0.02 |
ENSRNOT00000024178
|
Mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr7_+_11992543 | 0.02 |
ENSRNOT00000024576
|
Abhd17a
|
abhydrolase domain containing 17A |
| chr12_+_40553741 | 0.02 |
ENSRNOT00000057396
|
LOC100360238
|
rCG21419-like |
| chr14_+_43810119 | 0.02 |
ENSRNOT00000003327
|
Rbm47
|
RNA binding motif protein 47 |
| chrX_+_32495809 | 0.01 |
ENSRNOT00000020999
|
RGD1565844
|
similar to RIKEN cDNA 1700045I19 |
| chr2_+_157316266 | 0.01 |
ENSRNOT00000015387
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr3_+_93968855 | 0.01 |
ENSRNOT00000014478
|
Fbxo3
|
F-box protein 3 |
| chr3_-_161322289 | 0.01 |
ENSRNOT00000022594
|
Pltp
|
phospholipid transfer protein |
| chr12_+_41281559 | 0.01 |
ENSRNOT00000049622
ENSRNOT00000057266 |
Oas1e
|
2'-5' oligoadenylate synthetase 1E |
| chr11_-_76804510 | 0.01 |
ENSRNOT00000002649
|
Ccdc50
|
coiled-coil domain containing 50 |
| chr8_-_48692295 | 0.01 |
ENSRNOT00000065456
|
Vps11
|
VPS11 CORVET/HOPS core subunit |
| chr2_-_210738378 | 0.01 |
ENSRNOT00000025746
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr7_-_12918173 | 0.01 |
ENSRNOT00000011010
|
Tpgs1
|
tubulin polyglutamylase complex subunit 1 |
| chr11_+_87220618 | 0.01 |
ENSRNOT00000041975
|
Gsc2
|
goosecoid homeobox 2 |
| chr14_+_21041621 | 0.01 |
ENSRNOT00000086571
|
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr1_-_220771181 | 0.01 |
ENSRNOT00000027811
|
Sart1
|
squamous cell carcinoma antigen recognized by T cells 1 |
| chr3_-_14386118 | 0.01 |
ENSRNOT00000025649
|
Rab14
|
RAB14, member RAS oncogene family |
| chr4_+_158224000 | 0.01 |
ENSRNOT00000084240
ENSRNOT00000078495 |
Ano2
|
anoctamin 2 |
| chr13_-_77959089 | 0.01 |
ENSRNOT00000003586
|
Cacybp
|
calcyclin binding protein |
| chr8_+_21905865 | 0.01 |
ENSRNOT00000027973
|
Ppan
|
peter pan homolog (Drosophila) |
| chrX_+_63343546 | 0.01 |
ENSRNOT00000076315
|
Klhl15
|
kelch-like family member 15 |
| chr7_+_121480723 | 0.01 |
ENSRNOT00000065304
|
Atf4
|
activating transcription factor 4 |
| chr3_+_171743929 | 0.01 |
ENSRNOT00000079113
ENSRNOT00000064400 |
Rab22a
|
RAB22A, member RAS oncogene family |
| chrX_-_10772633 | 0.01 |
ENSRNOT00000035153
|
Ube2q2l
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr7_-_130347587 | 0.01 |
ENSRNOT00000040541
|
Tymp
|
thymidine phosphorylase |
| chr7_-_122967178 | 0.01 |
ENSRNOT00000052098
|
Rangap1
|
RAN GTPase activating protein 1 |
| chr13_-_93746994 | 0.01 |
ENSRNOT00000005072
|
Opn3
|
opsin 3 |
| chr18_-_27725694 | 0.01 |
ENSRNOT00000026398
|
Etf1
|
eukaryotic translation termination factor 1 |
| chr1_-_216703534 | 0.01 |
ENSRNOT00000027972
|
Phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chrX_+_29525256 | 0.01 |
ENSRNOT00000050018
|
Rab9a
|
RAB9A, member RAS oncogene family |
| chr18_+_29110242 | 0.01 |
ENSRNOT00000025756
|
Pura
|
purine rich element binding protein A |
| chr3_+_161343883 | 0.01 |
ENSRNOT00000023202
|
Pcif1
|
PDX1 C-terminal inhibiting factor 1 |
| chr12_+_10254951 | 0.01 |
ENSRNOT00000061129
|
Gpr12
|
G protein-coupled receptor 12 |
| chr20_+_46250363 | 0.01 |
ENSRNOT00000076522
ENSRNOT00000000334 |
Cd164
|
CD164 molecule |
| chr1_+_153589471 | 0.01 |
ENSRNOT00000023205
|
Fzd4
|
frizzled class receptor 4 |
| chr8_+_82257849 | 0.01 |
ENSRNOT00000011963
ENSRNOT00000075708 |
Gnb5
|
G protein subunit beta 5 |
| chr15_-_42751330 | 0.01 |
ENSRNOT00000066478
|
Adam2
|
ADAM metallopeptidase domain 2 |
| chr3_+_47439076 | 0.01 |
ENSRNOT00000011794
|
Tank
|
TRAF family member-associated NFKB activator |
| chr5_-_59292944 | 0.01 |
ENSRNOT00000051234
|
Olr838
|
olfactory receptor 838 |
| chr1_+_213766758 | 0.01 |
ENSRNOT00000005645
|
Ifitm1
|
interferon induced transmembrane protein 1 |
| chr5_+_48047913 | 0.01 |
ENSRNOT00000065926
|
Lyrm2
|
LYR motif containing 2 |
| chr9_-_99299715 | 0.01 |
ENSRNOT00000027622
|
Hdac4
|
histone deacetylase 4 |
| chr16_-_7007051 | 0.01 |
ENSRNOT00000023984
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr7_-_119623072 | 0.01 |
ENSRNOT00000050511
|
Tst
|
thiosulfate sulfurtransferase |
| chrX_-_4945944 | 0.01 |
ENSRNOT00000077238
|
Kdm6a
|
lysine demethylase 6A |
| chr1_+_143036218 | 0.01 |
ENSRNOT00000025797
|
Pde8a
|
phosphodiesterase 8A |
| chr16_+_20426566 | 0.00 |
ENSRNOT00000026225
|
Ifi30
|
IFI30, lysosomal thiol reductase |
| chr18_+_31444472 | 0.00 |
ENSRNOT00000075159
|
Rnf14
|
ring finger protein 14 |
| chr1_+_56982821 | 0.00 |
ENSRNOT00000059845
|
Ermard
|
ER membrane-associated RNA degradation |
| chr1_+_142136452 | 0.00 |
ENSRNOT00000016445
|
Hddc3
|
HD domain containing 3 |
| chr19_+_38846201 | 0.00 |
ENSRNOT00000030768
ENSRNOT00000063977 |
Tango6
|
transport and golgi organization 6 homolog |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.0 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.0 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.0 | GO:2000688 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 0.0 | 0.1 | GO:0044206 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) pyrimidine ribonucleoside catabolic process(GO:0046133) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0032426 | stereocilium tip(GO:0032426) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.0 | 0.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0005111 | type 1 fibroblast growth factor receptor binding(GO:0005105) type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.0 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |