Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
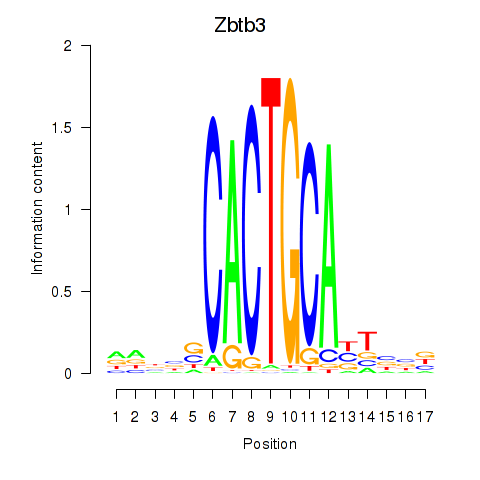
Results for Zbtb3
Z-value: 0.58
Transcription factors associated with Zbtb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb3
|
ENSRNOG00000019461 | zinc finger and BTB domain containing 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb3 | rn6_v1_chr1_+_224998172_224998172 | 0.85 | 6.9e-02 | Click! |
Activity profile of Zbtb3 motif
Sorted Z-values of Zbtb3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_71210301 | 0.39 |
ENSRNOT00000006504
|
Fam171b
|
family with sequence similarity 171, member B |
| chr16_-_84939323 | 0.35 |
ENSRNOT00000022742
|
Myo16
|
myosin XVI |
| chr1_+_226435979 | 0.34 |
ENSRNOT00000048704
ENSRNOT00000036232 ENSRNOT00000035576 ENSRNOT00000036180 ENSRNOT00000036168 ENSRNOT00000047964 ENSRNOT00000036283 ENSRNOT00000007429 |
Syt7
|
synaptotagmin 7 |
| chr1_-_261446570 | 0.23 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr7_-_113937941 | 0.20 |
ENSRNOT00000012408
|
Kcnk9
|
potassium two pore domain channel subfamily K member 9 |
| chr3_+_161433410 | 0.20 |
ENSRNOT00000024657
|
Slc12a5
|
solute carrier family 12 member 5 |
| chrX_+_65040934 | 0.20 |
ENSRNOT00000044006
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr1_-_65597899 | 0.19 |
ENSRNOT00000046708
|
Zfp446
|
zinc finger protein 446 |
| chr11_+_69739384 | 0.19 |
ENSRNOT00000016340
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr14_+_86652365 | 0.18 |
ENSRNOT00000077428
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chrX_+_31140960 | 0.17 |
ENSRNOT00000084395
ENSRNOT00000004494 |
Mospd2
|
motile sperm domain containing 2 |
| chr3_+_142739781 | 0.17 |
ENSRNOT00000006181
|
Sstr4
|
somatostatin receptor 4 |
| chr17_+_43632397 | 0.17 |
ENSRNOT00000013790
|
Hist1h2ah
|
histone cluster 1, H2ah |
| chr9_+_100885051 | 0.17 |
ENSRNOT00000082987
|
AC109427.1
|
|
| chr12_-_15996958 | 0.16 |
ENSRNOT00000039726
|
Amz1
|
archaelysin family metallopeptidase 1 |
| chr7_+_143749221 | 0.15 |
ENSRNOT00000014807
|
Igfbp6
|
insulin-like growth factor binding protein 6 |
| chr18_+_30435119 | 0.14 |
ENSRNOT00000027190
|
Pcdhb8
|
protocadherin beta 8 |
| chr13_-_52266100 | 0.14 |
ENSRNOT00000087532
|
AC096239.3
|
|
| chr7_-_12598183 | 0.13 |
ENSRNOT00000089009
|
Arid3a
|
AT-rich interaction domain 3A |
| chr7_-_12598370 | 0.13 |
ENSRNOT00000026708
|
Arid3a
|
AT-rich interaction domain 3A |
| chr20_+_28572242 | 0.13 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr4_-_176026133 | 0.13 |
ENSRNOT00000043374
ENSRNOT00000046598 |
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr2_-_210943620 | 0.13 |
ENSRNOT00000026750
|
Gpr61
|
G protein-coupled receptor 61 |
| chr10_+_14547172 | 0.12 |
ENSRNOT00000092043
|
Unkl
|
unkempt family like zinc finger |
| chr7_+_18510354 | 0.12 |
ENSRNOT00000060002
|
Pram1
|
PML-RARA regulated adaptor molecule 1 |
| chr6_+_119519714 | 0.12 |
ENSRNOT00000004955
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr10_-_103685844 | 0.11 |
ENSRNOT00000064284
|
Cd300lf
|
Cd300 molecule-like family member F |
| chr20_+_13067242 | 0.11 |
ENSRNOT00000083878
|
Dip2a
|
disco-interacting protein 2 homolog A |
| chr13_+_49850189 | 0.11 |
ENSRNOT00000042125
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chrX_+_75792931 | 0.11 |
ENSRNOT00000037575
|
Magee2
|
MAGE family member E2 |
| chr11_-_81444375 | 0.11 |
ENSRNOT00000058479
ENSRNOT00000078131 ENSRNOT00000080949 ENSRNOT00000080562 ENSRNOT00000084867 |
Kng1
|
kininogen 1 |
| chr10_+_35343189 | 0.10 |
ENSRNOT00000083688
|
Mapk9
|
mitogen-activated protein kinase 9 |
| chr4_-_100303047 | 0.10 |
ENSRNOT00000018170
ENSRNOT00000084782 |
Mat2a
|
methionine adenosyltransferase 2A |
| chr2_+_186980793 | 0.10 |
ENSRNOT00000091336
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr4_+_113247795 | 0.09 |
ENSRNOT00000007984
|
Tacr1
|
tachykinin receptor 1 |
| chr5_+_139963002 | 0.09 |
ENSRNOT00000048506
|
Col9a2
|
collagen type IX alpha 2 chain |
| chr3_+_48082935 | 0.09 |
ENSRNOT00000087711
ENSRNOT00000067545 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr10_+_13915214 | 0.09 |
ENSRNOT00000015092
|
Pkd1
|
polycystin 1, transient receptor potential channel interacting |
| chr18_+_87580424 | 0.09 |
ENSRNOT00000070984
|
Tmx3
|
thioredoxin-related transmembrane protein 3 |
| chr7_+_139762614 | 0.09 |
ENSRNOT00000031157
|
Ccdc184
|
coiled-coil domain containing 184 |
| chr3_-_94418711 | 0.09 |
ENSRNOT00000089554
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr3_-_29996865 | 0.09 |
ENSRNOT00000080382
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_86475096 | 0.09 |
ENSRNOT00000075247
ENSRNOT00000075062 |
LOC100909688
|
zinc finger protein 43-like |
| chr10_-_65502936 | 0.08 |
ENSRNOT00000089615
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr19_-_22632071 | 0.08 |
ENSRNOT00000077275
|
Gpt2
|
glutamic--pyruvic transaminase 2 |
| chr16_+_82184387 | 0.08 |
ENSRNOT00000089329
ENSRNOT00000068416 |
Tubgcp3
|
tubulin, gamma complex associated protein 3 |
| chr8_-_73194837 | 0.08 |
ENSRNOT00000024885
ENSRNOT00000081450 ENSRNOT00000064613 |
Tln2
|
talin 2 |
| chr6_+_1410507 | 0.08 |
ENSRNOT00000034919
|
Gpatch11
|
G patch domain containing 11 |
| chr15_+_41448064 | 0.08 |
ENSRNOT00000019551
|
Sacs
|
sacsin molecular chaperone |
| chr11_-_67702955 | 0.08 |
ENSRNOT00000084540
ENSRNOT00000078285 |
Kpna1
|
karyopherin subunit alpha 1 |
| chr20_+_29897594 | 0.08 |
ENSRNOT00000057752
|
Vsir
|
V-set immunoregulatory receptor |
| chr9_+_16702460 | 0.07 |
ENSRNOT00000061432
|
Ptk7
|
protein tyrosine kinase 7 |
| chr18_+_60639860 | 0.07 |
ENSRNOT00000083363
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr2_+_220298245 | 0.07 |
ENSRNOT00000022625
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr7_+_122979021 | 0.07 |
ENSRNOT00000085707
|
Zc3h7b
|
zinc finger CCCH-type containing 7B |
| chr17_+_15966234 | 0.06 |
ENSRNOT00000084304
|
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr1_+_48077033 | 0.06 |
ENSRNOT00000020100
|
Mas1
|
MAS1 proto-oncogene, G protein-coupled receptor |
| chr1_-_124038966 | 0.06 |
ENSRNOT00000082487
|
Chrna7
|
cholinergic receptor nicotinic alpha 7 subunit |
| chr11_-_71128299 | 0.06 |
ENSRNOT00000080568
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr5_-_166430254 | 0.06 |
ENSRNOT00000048914
|
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr1_+_67340809 | 0.06 |
ENSRNOT00000045266
|
LOC100362054
|
mCG114696-like |
| chr20_-_5927070 | 0.06 |
ENSRNOT00000059264
|
Slc26a8
|
solute carrier family 26 member 8 |
| chr3_-_44522930 | 0.06 |
ENSRNOT00000006963
|
Acvr1
|
activin A receptor type 1 |
| chr3_+_172692452 | 0.05 |
ENSRNOT00000077193
|
Zfp831
|
zinc finger protein 831 |
| chr15_-_3773340 | 0.05 |
ENSRNOT00000063831
|
LOC103689945
|
zinc finger SWIM domain-containing protein 8-like |
| chr5_-_62683964 | 0.05 |
ENSRNOT00000060005
|
Anks6
|
ankyrin repeat and sterile alpha motif domain containing 6 |
| chr15_+_5306459 | 0.05 |
ENSRNOT00000092079
|
LOC108348081
|
CD99 antigen-like protein 2 |
| chr10_+_40438356 | 0.05 |
ENSRNOT00000078910
|
Gm2a
|
GM2 ganglioside activator |
| chr6_-_79306443 | 0.05 |
ENSRNOT00000030706
|
Clec14a
|
C-type lectin domain family 14, member A |
| chr10_+_103934797 | 0.05 |
ENSRNOT00000035865
|
Cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr7_+_18409147 | 0.05 |
ENSRNOT00000086336
ENSRNOT00000011849 |
Adamts10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
| chr2_+_180012414 | 0.05 |
ENSRNOT00000082273
|
Pdgfc
|
platelet derived growth factor C |
| chr1_-_204817080 | 0.05 |
ENSRNOT00000077956
|
Fam53b
|
family with sequence similarity 53, member B |
| chr7_-_122963299 | 0.05 |
ENSRNOT00000090208
|
Rangap1
|
RAN GTPase activating protein 1 |
| chr10_-_35005287 | 0.04 |
ENSRNOT00000032386
|
LOC103689949
|
nedd4 binding protein 3 |
| chr8_+_44990014 | 0.04 |
ENSRNOT00000044608
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr9_-_88086488 | 0.04 |
ENSRNOT00000019579
|
Irs1
|
insulin receptor substrate 1 |
| chr20_+_3155652 | 0.04 |
ENSRNOT00000042882
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr10_+_45322248 | 0.04 |
ENSRNOT00000047268
|
Trim11
|
tripartite motif-containing 11 |
| chr1_-_124039196 | 0.04 |
ENSRNOT00000051883
|
Chrna7
|
cholinergic receptor nicotinic alpha 7 subunit |
| chr1_-_188713270 | 0.04 |
ENSRNOT00000082192
ENSRNOT00000065892 |
Gprc5b
|
G protein-coupled receptor, class C, group 5, member B |
| chr1_+_79931156 | 0.04 |
ENSRNOT00000019814
ENSRNOT00000084985 |
Sympk
|
symplekin |
| chr1_+_201256910 | 0.04 |
ENSRNOT00000090143
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr8_-_132911193 | 0.04 |
ENSRNOT00000087799
|
Fyco1
|
FYVE and coiled-coil domain containing 1 |
| chr4_+_70903253 | 0.04 |
ENSRNOT00000019720
|
Ephb6
|
Eph receptor B6 |
| chr3_+_67849966 | 0.04 |
ENSRNOT00000057826
|
Dusp19
|
dual specificity phosphatase 19 |
| chr5_-_160282810 | 0.03 |
ENSRNOT00000076160
ENSRNOT00000016352 |
Ddi2
|
DNA-damage inducible protein 2 |
| chr2_+_72006099 | 0.03 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr9_-_20195566 | 0.03 |
ENSRNOT00000015223
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chrX_-_75566481 | 0.03 |
ENSRNOT00000003714
|
Zdhhc15
|
zinc finger, DHHC-type containing 15 |
| chr14_+_79540235 | 0.03 |
ENSRNOT00000089334
|
Sorcs2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr17_+_30670171 | 0.03 |
ENSRNOT00000022369
|
Fam217a
|
family with sequence similarity 217, member A |
| chr1_+_214664537 | 0.03 |
ENSRNOT00000075002
|
AABR07006030.1
|
|
| chr15_+_43582020 | 0.03 |
ENSRNOT00000013001
|
Pnma2
|
paraneoplastic Ma antigen 2 |
| chr3_-_176666282 | 0.03 |
ENSRNOT00000016947
|
Eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr5_+_128215711 | 0.03 |
ENSRNOT00000011670
|
Cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr2_+_18392142 | 0.02 |
ENSRNOT00000043196
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr13_-_50761306 | 0.02 |
ENSRNOT00000076610
ENSRNOT00000004241 |
Prelp
|
proline and arginine rich end leucine rich repeat protein |
| chr19_-_10358695 | 0.02 |
ENSRNOT00000019770
|
Katnb1
|
katanin regulatory subunit B1 |
| chr8_+_31497639 | 0.02 |
ENSRNOT00000020041
|
Snx19
|
sorting nexin 19 |
| chr2_+_198231291 | 0.02 |
ENSRNOT00000078288
ENSRNOT00000046739 |
Otud7b
|
OTU deubiquitinase 7B |
| chr10_-_37099004 | 0.02 |
ENSRNOT00000075465
|
N4bp3
|
Nedd4 binding protein 3 |
| chr1_+_197128411 | 0.02 |
ENSRNOT00000083248
|
LOC361646
|
similar to K04F10.2 |
| chr8_-_122402127 | 0.02 |
ENSRNOT00000074238
ENSRNOT00000076548 |
Crtapl1
|
cartilage associated protein-like 1 |
| chr18_-_16543992 | 0.02 |
ENSRNOT00000036306
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr9_+_93799278 | 0.02 |
ENSRNOT00000084590
|
Dis3l2
|
DIS3-like 3'-5' exoribonuclease 2 |
| chr8_-_128622314 | 0.02 |
ENSRNOT00000084950
ENSRNOT00000024408 |
Gorasp1
|
golgi reassembly stacking protein 1 |
| chr5_-_173339890 | 0.02 |
ENSRNOT00000067704
ENSRNOT00000078453 |
Pusl1
|
pseudouridylate synthase-like 1 |
| chr1_+_265904566 | 0.02 |
ENSRNOT00000086041
ENSRNOT00000036156 |
Gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr16_+_50016857 | 0.02 |
ENSRNOT00000035781
|
Tlr3
|
toll-like receptor 3 |
| chr4_+_2711385 | 0.02 |
ENSRNOT00000035996
|
Dnajb6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr15_+_39945095 | 0.01 |
ENSRNOT00000016826
|
Shisa2
|
shisa family member 2 |
| chr8_-_22929294 | 0.01 |
ENSRNOT00000015609
|
Rab3d
|
RAB3D, member RAS oncogene family |
| chr20_+_31313018 | 0.01 |
ENSRNOT00000036719
|
Tysnd1
|
trypsin domain containing 1 |
| chr16_-_7588841 | 0.01 |
ENSRNOT00000084645
|
Mettl6
|
methyltransferase like 6 |
| chr2_-_30458542 | 0.01 |
ENSRNOT00000072926
|
Naip6
|
NLR family, apoptosis inhibitory protein 6 |
| chr20_-_45053640 | 0.01 |
ENSRNOT00000072256
|
RGD1561777
|
similar to Na+ dependent glucose transporter 1 |
| chr1_+_273854248 | 0.01 |
ENSRNOT00000044827
ENSRNOT00000017600 ENSRNOT00000086496 |
Add3
|
adducin 3 |
| chr6_-_1410339 | 0.01 |
ENSRNOT00000035905
|
Heatr5b
|
HEAT repeat containing 5B |
| chr2_-_27364906 | 0.01 |
ENSRNOT00000078639
|
Polk
|
DNA polymerase kappa |
| chr1_-_133503194 | 0.01 |
ENSRNOT00000077049
|
Mctp2
|
multiple C2 and transmembrane domain containing 2 |
| chr1_+_201055644 | 0.01 |
ENSRNOT00000054937
ENSRNOT00000047161 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr1_+_100830064 | 0.01 |
ENSRNOT00000027320
|
Il4i1
|
interleukin 4 induced 1 |
| chr10_-_91448477 | 0.01 |
ENSRNOT00000038836
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr8_-_39723927 | 0.00 |
ENSRNOT00000089662
|
Slc37a2
|
solute carrier family 37 member 2 |
| chr7_-_11783849 | 0.00 |
ENSRNOT00000026202
|
Sf3a2
|
splicing factor 3a, subunit 2 |
| chr4_-_77706994 | 0.00 |
ENSRNOT00000038517
|
Zfp777
|
zinc finger protein 777 |
| chr6_+_64252020 | 0.00 |
ENSRNOT00000047296
ENSRNOT00000082105 |
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr2_+_218951141 | 0.00 |
ENSRNOT00000091001
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr2_+_44096061 | 0.00 |
ENSRNOT00000018438
|
Ankrd55
|
ankyrin repeat domain 55 |
| chr1_-_219853329 | 0.00 |
ENSRNOT00000026169
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.2 | GO:2000040 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.2 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:1902214 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.2 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.0 | 0.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) metanephric proximal tubule development(GO:0072237) |
| 0.0 | 0.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.1 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.0 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) |
| 0.0 | 0.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.0 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.0 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.0 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.0 | 0.1 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |