Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
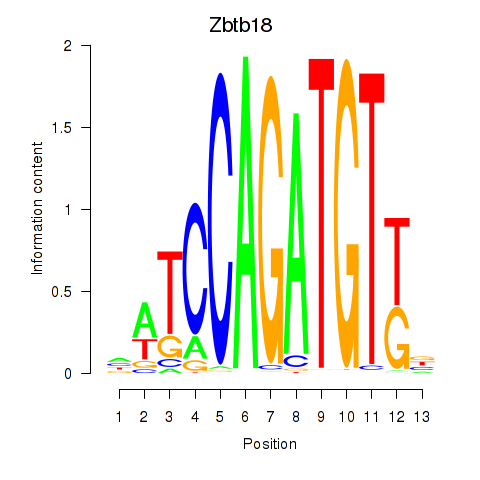
Results for Zbtb18
Z-value: 0.54
Transcription factors associated with Zbtb18
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp238 | rn6_v1_chr13_+_95589668_95589668 | -0.14 | 8.2e-01 | Click! |
Activity profile of Zbtb18 motif
Sorted Z-values of Zbtb18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_183674522 | 0.22 |
ENSRNOT00000014433
|
Tmem154
|
transmembrane protein 154 |
| chr1_-_52544450 | 0.19 |
ENSRNOT00000043474
|
Pde10a
|
phosphodiesterase 10A |
| chr4_-_132171153 | 0.18 |
ENSRNOT00000015058
ENSRNOT00000015075 |
Prok2
|
prokineticin 2 |
| chr10_+_2074995 | 0.17 |
ENSRNOT00000041674
|
RGD1562055
|
similar to ribosomal protein L31 |
| chr3_-_120306551 | 0.14 |
ENSRNOT00000021082
|
Mall
|
mal, T-cell differentiation protein-like |
| chr10_-_56962161 | 0.14 |
ENSRNOT00000026038
|
Alox15
|
arachidonate 15-lipoxygenase |
| chr1_-_101388125 | 0.13 |
ENSRNOT00000028176
ENSRNOT00000049957 |
Snrnp70
|
small nuclear ribonucleoprotein U1 subunit 70 |
| chr2_+_240021152 | 0.13 |
ENSRNOT00000012473
|
Tacr3
|
tachykinin receptor 3 |
| chr12_+_22641104 | 0.12 |
ENSRNOT00000001916
|
Serpine1
|
serpin family E member 1 |
| chr17_+_36690249 | 0.12 |
ENSRNOT00000089476
ENSRNOT00000082140 |
LOC688583
|
similar to High mobility group protein 4 (HMG-4) (High mobility group protein 2a) (HMG-2a) |
| chr11_+_89008008 | 0.12 |
ENSRNOT00000074586
|
Cebpd
|
CCAAT/enhancer binding protein delta |
| chr10_+_90930010 | 0.12 |
ENSRNOT00000003765
|
Higd1b
|
HIG1 hypoxia inducible domain family, member 1B |
| chr7_+_142905758 | 0.11 |
ENSRNOT00000078663
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_-_80744831 | 0.11 |
ENSRNOT00000025913
|
Bcl3
|
B-cell CLL/lymphoma 3 |
| chrX_-_106607352 | 0.11 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chr19_+_25391700 | 0.11 |
ENSRNOT00000011163
|
LOC100359539
|
ribonucleotide reductase M2 polypeptide |
| chr16_-_79973735 | 0.11 |
ENSRNOT00000057845
ENSRNOT00000086896 |
Dlgap2
|
DLG associated protein 2 |
| chr6_+_106039991 | 0.10 |
ENSRNOT00000088917
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr16_-_20148867 | 0.10 |
ENSRNOT00000040464
|
Fcho1
|
FCH domain only 1 |
| chr19_+_9024451 | 0.10 |
ENSRNOT00000073744
|
LOC100910721
|
60S ribosomal protein L26-like |
| chr9_-_60923706 | 0.10 |
ENSRNOT00000047517
|
AABR07067749.1
|
|
| chr9_-_79898912 | 0.10 |
ENSRNOT00000022076
|
March4
|
membrane associated ring-CH-type finger 4 |
| chr4_+_33638709 | 0.10 |
ENSRNOT00000009888
ENSRNOT00000034719 ENSRNOT00000052333 |
Tac1
|
tachykinin, precursor 1 |
| chr4_+_2055615 | 0.10 |
ENSRNOT00000007646
|
Rnf32
|
ring finger protein 32 |
| chr1_-_222177421 | 0.10 |
ENSRNOT00000078393
|
Esrra
|
estrogen related receptor, alpha |
| chr18_+_30895831 | 0.10 |
ENSRNOT00000026985
|
Pcdhga10
|
protocadherin gamma subfamily A, 10 |
| chr5_+_159845774 | 0.10 |
ENSRNOT00000012328
|
Epha2
|
Eph receptor A2 |
| chr11_+_45888221 | 0.09 |
ENSRNOT00000071932
ENSRNOT00000066008 ENSRNOT00000060802 |
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr17_+_43458553 | 0.09 |
ENSRNOT00000088939
|
Slc17a4
|
solute carrier family 17, member 4 |
| chr20_-_6548179 | 0.09 |
ENSRNOT00000093711
|
Lemd2
|
LEM domain containing 2 |
| chr8_+_63286773 | 0.09 |
ENSRNOT00000033397
|
RGD1562618
|
similar to RIKEN cDNA 6030419C18 gene |
| chr1_-_81450094 | 0.09 |
ENSRNOT00000035356
|
Zfp575
|
zinc finger protein 575 |
| chr4_-_157408176 | 0.09 |
ENSRNOT00000021915
|
Cd4
|
Cd4 molecule |
| chr1_+_80426918 | 0.09 |
ENSRNOT00000067272
|
Nkpd1
|
NTPase, KAP family P-loop domain containing 1 |
| chr14_+_37435654 | 0.09 |
ENSRNOT00000002991
|
Ociad2
|
OCIA domain containing 2 |
| chr4_-_79015660 | 0.09 |
ENSRNOT00000068441
|
LOC100910678
|
coiled-coil domain-containing protein 72-like |
| chr7_-_18577325 | 0.09 |
ENSRNOT00000084308
ENSRNOT00000090849 |
March2
|
membrane associated ring-CH-type finger 2 |
| chr6_+_106153045 | 0.09 |
ENSRNOT00000092065
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr4_+_170347410 | 0.09 |
ENSRNOT00000040508
|
LOC500350
|
LRRGT00139 |
| chr15_-_14622587 | 0.08 |
ENSRNOT00000086502
|
Synpr
|
synaptoporin |
| chr3_+_171832500 | 0.08 |
ENSRNOT00000007554
|
Vapb
|
VAMP associated protein B and C |
| chr4_+_179398621 | 0.08 |
ENSRNOT00000049474
ENSRNOT00000067506 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr1_+_192613372 | 0.08 |
ENSRNOT00000016632
|
Cacng3
|
calcium voltage-gated channel auxiliary subunit gamma 3 |
| chr13_-_91228901 | 0.08 |
ENSRNOT00000071728
ENSRNOT00000073643 ENSRNOT00000071897 |
LOC100911825
|
low affinity immunoglobulin gamma Fc region receptor III-like |
| chr1_+_72545596 | 0.08 |
ENSRNOT00000022698
|
Shisa7
|
shisa family member 7 |
| chr13_+_83073866 | 0.08 |
ENSRNOT00000075996
|
Dpt
|
dermatopontin |
| chr20_-_5618254 | 0.08 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chr8_-_84506328 | 0.08 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chrX_-_26376467 | 0.08 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr1_+_167538744 | 0.08 |
ENSRNOT00000093070
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr6_-_136379348 | 0.08 |
ENSRNOT00000016208
|
Xrcc3
|
X-ray repair cross complementing 3 |
| chr13_-_72063347 | 0.07 |
ENSRNOT00000090544
ENSRNOT00000003869 |
Cacna1e
|
calcium voltage-gated channel subunit alpha1 E |
| chr3_+_162346490 | 0.07 |
ENSRNOT00000087530
|
Eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr18_+_30909490 | 0.07 |
ENSRNOT00000026967
|
Pcdhgb8
|
protocadherin gamma subfamily B, 8 |
| chr1_+_261497059 | 0.07 |
ENSRNOT00000020390
|
Golga7b
|
golgin A7 family, member B |
| chr2_+_180012414 | 0.07 |
ENSRNOT00000082273
|
Pdgfc
|
platelet derived growth factor C |
| chr12_-_18531161 | 0.07 |
ENSRNOT00000037829
|
Akap17a
|
A-kinase anchoring protein 17A |
| chr14_+_12218553 | 0.07 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr10_-_106883423 | 0.07 |
ENSRNOT00000071406
|
LOC688286
|
similar to threonine aldolase 1 |
| chr2_+_187344056 | 0.07 |
ENSRNOT00000025314
|
Nes
|
nestin |
| chr1_+_87045335 | 0.07 |
ENSRNOT00000084393
|
Lgals7
|
galectin 7 |
| chrX_+_22788660 | 0.07 |
ENSRNOT00000071886
|
AABR07037510.1
|
|
| chr4_-_117268178 | 0.07 |
ENSRNOT00000043201
ENSRNOT00000084049 |
Fbxo41
|
F-box protein 41 |
| chr8_-_70436028 | 0.07 |
ENSRNOT00000077152
|
Slc24a1
|
solute carrier family 24 member 1 |
| chr1_-_167165265 | 0.07 |
ENSRNOT00000027321
|
Rnf121
|
ring finger protein 121 |
| chr1_+_100299626 | 0.07 |
ENSRNOT00000092327
ENSRNOT00000044257 |
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr6_-_127248372 | 0.07 |
ENSRNOT00000085517
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chrX_+_68782872 | 0.07 |
ENSRNOT00000075995
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr3_-_147809919 | 0.07 |
ENSRNOT00000085849
|
Rbck1
|
RANBP2-type and C3HC4-type zinc finger containing 1 |
| chr3_-_123698072 | 0.07 |
ENSRNOT00000077819
|
RGD1311739
|
similar to RIKEN cDNA 1700037H04 |
| chr8_-_104155775 | 0.07 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chr5_-_155381732 | 0.07 |
ENSRNOT00000033670
|
Zbtb40
|
zinc finger and BTB domain containing 40 |
| chr20_+_47779056 | 0.07 |
ENSRNOT00000090015
|
AC132752.1
|
|
| chr7_-_118792625 | 0.07 |
ENSRNOT00000007398
|
Myh9
|
myosin, heavy chain 9 |
| chr6_+_48866601 | 0.07 |
ENSRNOT00000077321
ENSRNOT00000079891 |
Pxdn
|
peroxidasin |
| chr3_-_143063983 | 0.07 |
ENSRNOT00000006329
|
Napb
|
NSF attachment protein beta |
| chr1_-_199177365 | 0.06 |
ENSRNOT00000041840
|
Ctf2
|
cardiotrophin 2 |
| chr11_-_88177448 | 0.06 |
ENSRNOT00000085475
|
Ypel1
|
yippee-like 1 |
| chr5_-_172424081 | 0.06 |
ENSRNOT00000019096
|
Plch2
|
phospholipase C, eta 2 |
| chr2_+_193892589 | 0.06 |
ENSRNOT00000035240
|
S100a10
|
S100 calcium binding protein A10 |
| chr7_-_8060373 | 0.06 |
ENSRNOT00000071699
|
Olfr798
|
olfactory receptor 798 |
| chr11_-_72934823 | 0.06 |
ENSRNOT00000075353
|
Pigz
|
phosphatidylinositol glycan anchor biosynthesis, class Z |
| chr3_-_160573978 | 0.06 |
ENSRNOT00000018386
|
Wfdc5
|
WAP four-disulfide core domain 5 |
| chr3_+_154395187 | 0.06 |
ENSRNOT00000050810
|
Vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr2_-_199771896 | 0.06 |
ENSRNOT00000043937
|
Chd1l
|
chromodomain helicase DNA binding protein 1-like |
| chr19_+_9436210 | 0.06 |
ENSRNOT00000074845
|
LOC100910721
|
60S ribosomal protein L26-like |
| chr10_-_62254287 | 0.06 |
ENSRNOT00000004313
|
Serpinf1
|
serpin family F member 1 |
| chr3_-_165477771 | 0.06 |
ENSRNOT00000071672
|
Atp9a
|
ATPase phospholipid transporting 9A (putative) |
| chr7_+_27089278 | 0.06 |
ENSRNOT00000082398
|
Nfyb
|
nuclear transcription factor Y subunit beta |
| chr1_+_211351350 | 0.06 |
ENSRNOT00000082751
ENSRNOT00000030461 |
Jakmip3
|
janus kinase and microtubule interacting protein 3 |
| chr11_-_72086743 | 0.06 |
ENSRNOT00000072173
|
Pigz
|
phosphatidylinositol glycan anchor biosynthesis, class Z |
| chr14_+_2311122 | 0.06 |
ENSRNOT00000037459
|
Mfsd7
|
major facilitator superfamily domain containing 7 |
| chr9_+_12114977 | 0.06 |
ENSRNOT00000073673
|
AABR07066648.1
|
|
| chrX_-_39956841 | 0.06 |
ENSRNOT00000050708
|
Klhl34
|
kelch-like family member 34 |
| chr2_+_252452269 | 0.06 |
ENSRNOT00000021970
|
Uox
|
urate oxidase |
| chr6_-_45669148 | 0.05 |
ENSRNOT00000010092
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr17_-_85141210 | 0.05 |
ENSRNOT00000000162
|
Dnajc1
|
DnaJ heat shock protein family (Hsp40) member C1 |
| chr5_-_164709441 | 0.05 |
ENSRNOT00000091132
|
LOC100911485
|
mitofusin-2-like |
| chr9_-_8632017 | 0.05 |
ENSRNOT00000043269
|
AABR07066416.1
|
|
| chr4_+_120671489 | 0.05 |
ENSRNOT00000029125
|
Mgll
|
monoglyceride lipase |
| chr15_+_57709043 | 0.05 |
ENSRNOT00000030579
|
Erich6b
|
glutamate-rich 6B |
| chr8_-_87282156 | 0.05 |
ENSRNOT00000087874
|
Filip1
|
filamin A interacting protein 1 |
| chr3_+_11679530 | 0.05 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr5_-_164585634 | 0.05 |
ENSRNOT00000065985
|
Mfn2
|
mitofusin 2 |
| chr1_+_236031988 | 0.05 |
ENSRNOT00000016164
|
Pcsk5
|
proprotein convertase subtilisin/kexin type 5 |
| chr9_+_111249351 | 0.05 |
ENSRNOT00000076729
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr12_+_41543696 | 0.05 |
ENSRNOT00000079639
|
Tpcn1
|
two pore segment channel 1 |
| chr7_-_130128589 | 0.05 |
ENSRNOT00000079777
ENSRNOT00000009325 |
Mapk11
|
mitogen-activated protein kinase 11 |
| chr1_-_82580761 | 0.05 |
ENSRNOT00000028131
ENSRNOT00000028132 |
Axl
|
Axl receptor tyrosine kinase |
| chr8_-_33121002 | 0.05 |
ENSRNOT00000047211
|
LOC100362981
|
LRRGT00010-like |
| chr3_-_95418679 | 0.05 |
ENSRNOT00000018074
|
Rcn1
|
reticulocalbin 1 |
| chr4_+_172942020 | 0.05 |
ENSRNOT00000072450
|
Lmo3
|
LIM domain only 3 |
| chr15_+_37806836 | 0.05 |
ENSRNOT00000076285
|
Il17d
|
interleukin 17D |
| chr9_-_100479868 | 0.05 |
ENSRNOT00000022748
|
Pask
|
PAS domain containing serine/threonine kinase |
| chr19_-_25790077 | 0.05 |
ENSRNOT00000091670
|
Nacc1
|
nucleus accumbens associated 1 |
| chr4_+_170958196 | 0.05 |
ENSRNOT00000007905
|
Pde6h
|
phosphodiesterase 6H |
| chr15_-_34469746 | 0.05 |
ENSRNOT00000027719
|
Adcy4
|
adenylate cyclase 4 |
| chr17_-_54714914 | 0.05 |
ENSRNOT00000024336
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr16_+_11599753 | 0.05 |
ENSRNOT00000079833
|
Grid1
|
glutamate ionotropic receptor delta type subunit 1 |
| chrX_-_107442878 | 0.05 |
ENSRNOT00000052302
|
Glra4
|
glycine receptor, alpha 4 |
| chr3_+_122816924 | 0.05 |
ENSRNOT00000045604
|
RGD1561317
|
similar to ribosomal protein L31 |
| chr2_+_143895982 | 0.05 |
ENSRNOT00000080026
|
Supt20h
|
SPT20 homolog, SAGA complex component |
| chrX_-_63292092 | 0.05 |
ENSRNOT00000081124
|
Eif2s3
|
eukaryotic translation initiation factor 2 subunit gamma |
| chr2_-_187820952 | 0.05 |
ENSRNOT00000092762
|
Sema4a
|
semaphorin 4A |
| chr16_-_10529061 | 0.05 |
ENSRNOT00000086215
|
Gprin2
|
G protein regulated inducer of neurite outgrowth 2 |
| chr1_+_274688580 | 0.05 |
ENSRNOT00000020580
|
Shoc2
|
SHOC2 leucine-rich repeat scaffold protein |
| chr10_-_90151042 | 0.05 |
ENSRNOT00000055187
|
Hdac5
|
histone deacetylase 5 |
| chrX_-_116638508 | 0.05 |
ENSRNOT00000030400
|
Lhfpl1
|
lipoma HMGIC fusion partner-like 1 |
| chr4_+_7257752 | 0.05 |
ENSRNOT00000088357
|
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr4_-_133951264 | 0.05 |
ENSRNOT00000090506
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr8_+_14060394 | 0.05 |
ENSRNOT00000014827
|
Smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr1_-_215284548 | 0.05 |
ENSRNOT00000090375
|
Cttn
|
cortactin |
| chr3_+_5624506 | 0.05 |
ENSRNOT00000036995
|
Adamtsl2
|
ADAMTS-like 2 |
| chr18_+_59479788 | 0.05 |
ENSRNOT00000015408
|
RGD1565894
|
similar to ribosomal protein L31 |
| chr17_+_76079720 | 0.05 |
ENSRNOT00000073933
|
Proser2
|
proline and serine rich 2 |
| chr7_-_15821927 | 0.05 |
ENSRNOT00000050658
|
LOC691422
|
similar to zinc finger protein 101 |
| chr10_-_52483325 | 0.05 |
ENSRNOT00000083485
|
AABR07029809.1
|
|
| chr19_+_14835822 | 0.05 |
ENSRNOT00000072804
|
1700007B14Rik
|
RIKEN cDNA 1700007B14 gene |
| chr2_+_143656793 | 0.05 |
ENSRNOT00000084527
ENSRNOT00000017453 |
Postn
|
periostin |
| chr3_+_80556668 | 0.04 |
ENSRNOT00000079118
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr1_-_88690534 | 0.04 |
ENSRNOT00000072570
|
Ovol3
|
ovo-like zinc finger 3 |
| chr10_+_89251370 | 0.04 |
ENSRNOT00000076820
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr10_-_35716260 | 0.04 |
ENSRNOT00000004308
ENSRNOT00000059255 |
Sqstm1
|
sequestosome 1 |
| chr10_+_17261541 | 0.04 |
ENSRNOT00000005478
|
Sh3pxd2b
|
SH3 and PX domains 2B |
| chr7_+_116632506 | 0.04 |
ENSRNOT00000009811
|
Gpihbp1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr20_+_3178250 | 0.04 |
ENSRNOT00000082116
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr16_-_21338771 | 0.04 |
ENSRNOT00000014265
|
Pbx4
|
PBX homeobox 4 |
| chr4_-_118534266 | 0.04 |
ENSRNOT00000081201
ENSRNOT00000024414 |
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr10_+_107455845 | 0.04 |
ENSRNOT00000004373
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr10_+_109446444 | 0.04 |
ENSRNOT00000066736
|
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr3_-_118427851 | 0.04 |
ENSRNOT00000055995
|
Fam227b
|
family with sequence similarity 227, member B |
| chr16_-_6245644 | 0.04 |
ENSRNOT00000040759
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr1_+_128199322 | 0.04 |
ENSRNOT00000075131
|
Lysmd4
|
LysM domain containing 4 |
| chr9_+_17225122 | 0.04 |
ENSRNOT00000068747
ENSRNOT00000080816 |
Rsph9
|
radial spoke head 9 homolog |
| chr4_-_119927705 | 0.04 |
ENSRNOT00000077624
|
Rab7a
|
RAB7A, member RAS oncogene family |
| chr13_-_89433815 | 0.04 |
ENSRNOT00000091541
|
Fcgr2b
|
Fc fragment of IgG receptor IIb |
| chr1_-_217631862 | 0.04 |
ENSRNOT00000078514
ENSRNOT00000079805 |
Cttn
|
cortactin |
| chr8_+_49737798 | 0.04 |
ENSRNOT00000022476
|
Dscaml1
|
DS cell adhesion molecule-like 1 |
| chr3_+_4233111 | 0.04 |
ENSRNOT00000042910
|
Lcn1
|
lipocalin 1 |
| chr12_-_40822159 | 0.04 |
ENSRNOT00000001826
|
Hectd4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr8_-_78233430 | 0.04 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr5_-_139864393 | 0.04 |
ENSRNOT00000030016
|
Zfp69
|
zinc finger protein 69 |
| chr6_+_107517668 | 0.04 |
ENSRNOT00000013753
|
Acot4
|
acyl-CoA thioesterase 4 |
| chr1_-_164659992 | 0.04 |
ENSRNOT00000024281
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr6_+_73553210 | 0.04 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr10_+_15672382 | 0.04 |
ENSRNOT00000027965
|
Rhbdf1
|
rhomboid 5 homolog 1 |
| chr13_+_53283855 | 0.04 |
ENSRNOT00000043590
|
RGD1564839
|
similar to ribosomal protein L31 |
| chr8_+_5768811 | 0.04 |
ENSRNOT00000013936
|
Mmp8
|
matrix metallopeptidase 8 |
| chr18_+_81569910 | 0.04 |
ENSRNOT00000020858
|
Fam69c
|
family with sequence similarity 69, member C |
| chr2_+_189629297 | 0.04 |
ENSRNOT00000049810
|
Dennd4b
|
DENN domain containing 4B |
| chr11_-_77593171 | 0.04 |
ENSRNOT00000002645
ENSRNOT00000043498 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr4_+_174810959 | 0.04 |
ENSRNOT00000035507
|
Aebp2
|
AE binding protein 2 |
| chr11_+_9642365 | 0.04 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr1_-_278042312 | 0.04 |
ENSRNOT00000018693
|
Ablim1
|
actin-binding LIM protein 1 |
| chr1_-_197568165 | 0.04 |
ENSRNOT00000079044
|
Xpo6
|
exportin 6 |
| chr8_-_105462141 | 0.04 |
ENSRNOT00000066731
ENSRNOT00000078760 |
Clstn2
|
calsyntenin 2 |
| chr2_-_210282352 | 0.04 |
ENSRNOT00000075653
|
Slc6a17
|
solute carrier family 6 member 17 |
| chr7_-_140172448 | 0.04 |
ENSRNOT00000077672
|
LOC103692976
|
cyclin-T1-like |
| chr10_-_103481153 | 0.04 |
ENSRNOT00000035639
|
Cd300lb
|
CD300 molecule-like family member b |
| chr7_+_123510804 | 0.04 |
ENSRNOT00000010491
|
Sept3
|
septin 3 |
| chr4_-_22192474 | 0.04 |
ENSRNOT00000043822
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr4_-_157274755 | 0.04 |
ENSRNOT00000088895
|
LOC100911672
|
atrophin-1-like |
| chr13_+_52413241 | 0.04 |
ENSRNOT00000035157
|
AABR07020999.1
|
|
| chr2_+_189997129 | 0.04 |
ENSRNOT00000015958
|
S100a4
|
S100 calcium-binding protein A4 |
| chrX_+_137934484 | 0.04 |
ENSRNOT00000049046
|
LOC317588
|
hypothetical protein LOC317588 |
| chr20_-_5107899 | 0.04 |
ENSRNOT00000038130
ENSRNOT00000086413 |
Ly6g5b
|
lymphocyte antigen 6 complex, locus G5B |
| chr9_+_27311886 | 0.04 |
ENSRNOT00000038486
|
RGD1562107
|
similar to class-alpha glutathione S-transferase |
| chr17_+_44522140 | 0.04 |
ENSRNOT00000080490
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr11_-_62451149 | 0.04 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr20_+_25990304 | 0.04 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr10_+_90342051 | 0.04 |
ENSRNOT00000028487
|
Rundc3a
|
RUN domain containing 3A |
| chr11_-_67702955 | 0.04 |
ENSRNOT00000084540
ENSRNOT00000078285 |
Kpna1
|
karyopherin subunit alpha 1 |
| chr1_-_163554839 | 0.04 |
ENSRNOT00000081509
|
Emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr7_-_139254551 | 0.04 |
ENSRNOT00000083182
|
Rapgef3
|
Rap guanine nucleotide exchange factor 3 |
| chr10_-_62664466 | 0.03 |
ENSRNOT00000078730
|
Git1
|
GIT ArfGAP 1 |
| chr10_+_45659143 | 0.03 |
ENSRNOT00000058327
|
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr12_-_20486276 | 0.03 |
ENSRNOT00000074057
|
LOC680910
|
similar to paired immunoglobin-like type 2 receptor beta |
| chr16_-_81880502 | 0.03 |
ENSRNOT00000079096
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr10_-_54467956 | 0.03 |
ENSRNOT00000065383
|
Usp43
|
ubiquitin specific peptidase 43 |
| chr1_+_214534284 | 0.03 |
ENSRNOT00000064254
|
Ap2a2
|
adaptor-related protein complex 2, alpha 2 subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb18
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000097 | response to resveratrol(GO:1904638) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 0.0 | 0.1 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.1 | GO:0045105 | intermediate filament polymerization or depolymerization(GO:0045105) |
| 0.0 | 0.1 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.0 | 0.0 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.0 | GO:0032759 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.0 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.0 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.0 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0002339 | B cell selection(GO:0002339) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.0 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.0 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.0 | GO:0015820 | leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.0 | GO:0032197 | transposition, RNA-mediated(GO:0032197) |
| 0.0 | 0.0 | GO:2000669 | neutrophil clearance(GO:0097350) negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.0 | 0.0 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.0 | 0.1 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.0 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0099571 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 0.0 | 0.0 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0031762 | follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0030619 | U1 snRNA binding(GO:0030619) U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.0 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |