Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
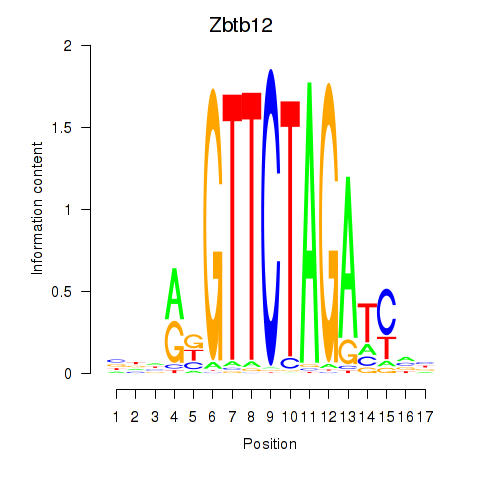
Results for Zbtb12
Z-value: 0.35
Transcription factors associated with Zbtb12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb12
|
ENSRNOG00000000418 | zinc finger and BTB domain containing 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb12 | rn6_v1_chr20_+_4572100_4572100 | 0.03 | 9.7e-01 | Click! |
Activity profile of Zbtb12 motif
Sorted Z-values of Zbtb12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_187447501 | 0.13 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr12_-_19254527 | 0.12 |
ENSRNOT00000089349
|
AABR07035541.3
|
|
| chr2_-_265300868 | 0.10 |
ENSRNOT00000066024
ENSRNOT00000016073 ENSRNOT00000033502 |
Lrrc7
|
leucine rich repeat containing 7 |
| chr13_+_71656651 | 0.08 |
ENSRNOT00000050365
|
Zfp648
|
zinc finger protein 648 |
| chrX_-_121731543 | 0.08 |
ENSRNOT00000018788
|
Klhl13
|
kelch-like family member 13 |
| chr12_+_47074200 | 0.07 |
ENSRNOT00000014910
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr1_+_81763614 | 0.07 |
ENSRNOT00000027254
|
Cd79a
|
CD79a molecule |
| chr1_-_266333105 | 0.06 |
ENSRNOT00000027093
|
Arl3
|
ADP ribosylation factor like GTPase 3 |
| chr19_-_37819789 | 0.06 |
ENSRNOT00000036702
|
Cenpt
|
centromere protein T |
| chr10_-_29026002 | 0.05 |
ENSRNOT00000005070
|
Pttg1
|
pituitary tumor-transforming 1 |
| chr1_-_141349881 | 0.05 |
ENSRNOT00000021109
|
Rhcg
|
Rh family, C glycoprotein |
| chr1_+_80135391 | 0.05 |
ENSRNOT00000021893
|
Gpr4
|
G protein-coupled receptor 4 |
| chrX_-_112328642 | 0.05 |
ENSRNOT00000083150
|
Psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr3_+_101899068 | 0.05 |
ENSRNOT00000079600
ENSRNOT00000006256 |
Muc15
|
mucin 15, cell surface associated |
| chr18_-_68983619 | 0.05 |
ENSRNOT00000014670
|
LOC361346
|
similar to chromosome 18 open reading frame 54 |
| chr13_-_97282299 | 0.04 |
ENSRNOT00000004244
|
Mixl1
|
Mix paired-like homeobox 1 |
| chr20_+_3178250 | 0.04 |
ENSRNOT00000082116
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr3_+_162357356 | 0.04 |
ENSRNOT00000030119
|
Eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr9_+_40817654 | 0.04 |
ENSRNOT00000037392
|
AABR07067339.1
|
|
| chr7_-_140580783 | 0.04 |
ENSRNOT00000087327
|
Dhh
|
desert hedgehog |
| chr3_-_123997277 | 0.04 |
ENSRNOT00000058238
|
RGD1564425
|
similar to Proteasome subunit beta type 3 (Proteasome theta chain) |
| chr1_-_78626284 | 0.04 |
ENSRNOT00000052299
|
LOC108349752
|
carcinoembryonic antigen-related cell adhesion molecule 3-like |
| chr15_-_39705208 | 0.04 |
ENSRNOT00000015412
|
Phf11
|
PHD finger protein 11 |
| chr3_+_100787449 | 0.04 |
ENSRNOT00000078543
|
Bdnf
|
brain-derived neurotrophic factor |
| chr3_-_165537940 | 0.04 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr4_+_7258176 | 0.04 |
ENSRNOT00000061992
|
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr7_+_123482255 | 0.04 |
ENSRNOT00000064487
|
LOC688613
|
hypothetical protein LOC688613 |
| chr19_-_26946044 | 0.04 |
ENSRNOT00000090756
|
AABR07043271.1
|
|
| chr15_-_61772516 | 0.03 |
ENSRNOT00000015605
ENSRNOT00000093399 |
Wbp4
|
WW domain binding protein 4 |
| chr10_+_99437436 | 0.03 |
ENSRNOT00000006254
|
Kcnj2
|
potassium voltage-gated channel subfamily J member 2 |
| chr2_+_113984824 | 0.03 |
ENSRNOT00000086399
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr18_+_68983545 | 0.03 |
ENSRNOT00000085317
|
Stard6
|
StAR-related lipid transfer domain containing 6 |
| chr8_+_39164916 | 0.03 |
ENSRNOT00000011309
|
Acrv1
|
acrosomal vesicle protein 1 |
| chr1_+_83003841 | 0.03 |
ENSRNOT00000057384
|
Ceacam4
|
carcinoembryonic antigen-related cell adhesion molecule 4 |
| chr6_+_104068547 | 0.03 |
ENSRNOT00000006460
|
Galnt16
|
polypeptide N-acetylgalactosaminyltransferase 16 |
| chr11_-_29710849 | 0.03 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr19_-_26243223 | 0.03 |
ENSRNOT00000037766
|
Zfp791
|
zinc finger protein 791 |
| chr8_-_73122740 | 0.03 |
ENSRNOT00000012026
|
Tln2
|
talin 2 |
| chr6_-_91518996 | 0.03 |
ENSRNOT00000005835
|
Pole2
|
DNA polymerase epsilon 2, accessory subunit |
| chr9_+_17041389 | 0.03 |
ENSRNOT00000025598
|
Abcc10
|
ATP binding cassette subfamily C member 10 |
| chr3_+_100786862 | 0.03 |
ENSRNOT00000080190
|
Bdnf
|
brain-derived neurotrophic factor |
| chr6_-_36940868 | 0.03 |
ENSRNOT00000006470
|
Gen1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr7_-_117722359 | 0.03 |
ENSRNOT00000019876
|
Cyhr1
|
cysteine and histidine rich 1 |
| chr1_+_103145939 | 0.03 |
ENSRNOT00000040529
|
AABR07003273.1
|
|
| chr19_+_26416818 | 0.03 |
ENSRNOT00000040473
|
AABR07043200.1
|
|
| chr3_+_55886695 | 0.02 |
ENSRNOT00000009396
|
Bbs5
|
Bardet-Biedl syndrome 5 |
| chr1_+_266333440 | 0.02 |
ENSRNOT00000071243
|
Sfxn2
|
sideroflexin 2 |
| chr9_-_14550625 | 0.02 |
ENSRNOT00000078227
|
Oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr11_-_64855353 | 0.02 |
ENSRNOT00000089625
|
Cd80
|
Cd80 molecule |
| chr6_+_129609074 | 0.02 |
ENSRNOT00000064957
|
Papola
|
poly (A) polymerase alpha |
| chr1_+_213597478 | 0.02 |
ENSRNOT00000017562
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr2_+_206314213 | 0.02 |
ENSRNOT00000056068
|
Bcl2l15
|
BCL2-like 15 |
| chr16_+_9907598 | 0.02 |
ENSRNOT00000027352
|
Ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr1_+_81643816 | 0.02 |
ENSRNOT00000027214
|
LOC103689942
|
carcinoembryonic antigen-related cell adhesion molecule 1-like |
| chr20_-_5805627 | 0.02 |
ENSRNOT00000085996
|
Clps
|
colipase |
| chr3_+_8928534 | 0.02 |
ENSRNOT00000088509
ENSRNOT00000065300 |
Miga2
|
mitoguardin 2 |
| chr6_+_129609375 | 0.02 |
ENSRNOT00000083732
|
Papola
|
poly (A) polymerase alpha |
| chr12_+_37829579 | 0.02 |
ENSRNOT00000084587
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr3_+_47677720 | 0.02 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
| chr3_-_23353425 | 0.02 |
ENSRNOT00000019703
|
Golga1
|
golgin A1 |
| chr1_+_198409360 | 0.02 |
ENSRNOT00000013691
ENSRNOT00000091295 |
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr4_-_51598771 | 0.01 |
ENSRNOT00000008325
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr1_-_216971183 | 0.01 |
ENSRNOT00000077911
|
Mrgpre
|
MAS related GPR family member E |
| chr19_-_26129619 | 0.01 |
ENSRNOT00000067600
|
Best2
|
bestrophin 2 |
| chrX_+_6430594 | 0.01 |
ENSRNOT00000044009
|
Maob
|
monoamine oxidase B |
| chr2_-_250862419 | 0.01 |
ENSRNOT00000017943
|
Clca4
|
chloride channel accessory 4 |
| chr5_-_58106706 | 0.01 |
ENSRNOT00000019561
|
Dctn3
|
dynactin subunit 3 |
| chr20_+_19325121 | 0.01 |
ENSRNOT00000058151
|
Phyhipl
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr10_+_90988732 | 0.01 |
ENSRNOT00000003892
|
FAM187A
|
family with sequence similarity 187, member A |
| chr1_-_225035687 | 0.01 |
ENSRNOT00000026539
|
Gng3
|
G protein subunit gamma 3 |
| chr9_-_61690956 | 0.01 |
ENSRNOT00000066589
|
Hspd1
|
heat shock protein family D member 1 |
| chr15_-_33775109 | 0.01 |
ENSRNOT00000033722
|
Jph4
|
junctophilin 4 |
| chr7_-_123586919 | 0.01 |
ENSRNOT00000011484
|
Ndufa6
|
NADH:ubiquinone oxidoreductase subunit A6 |
| chr7_-_117721955 | 0.01 |
ENSRNOT00000019853
|
Cyhr1
|
cysteine and histidine rich 1 |
| chr3_+_28416954 | 0.01 |
ENSRNOT00000043533
|
Kynu
|
kynureninase |
| chr1_+_280423079 | 0.01 |
ENSRNOT00000011983
|
Slc18a2
|
solute carrier family 18 member A2 |
| chr6_+_22296128 | 0.01 |
ENSRNOT00000087805
|
Dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr10_-_86509454 | 0.01 |
ENSRNOT00000009454
|
Ikzf3
|
IKAROS family zinc finger 3 |
| chr8_+_87205311 | 0.01 |
ENSRNOT00000014449
|
LOC100363289
|
LRRGT00022-like |
| chr3_-_113130058 | 0.01 |
ENSRNOT00000046391
ENSRNOT00000035889 |
Zscan29
|
zinc finger and SCAN domain containing 29 |
| chr1_+_189328246 | 0.01 |
ENSRNOT00000084260
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr10_-_5253336 | 0.01 |
ENSRNOT00000085310
|
Ciita
|
class II, major histocompatibility complex, transactivator |
| chr6_-_96202477 | 0.01 |
ENSRNOT00000046585
|
LOC688981
|
similar to 60S ribosomal protein L26 (Silica-induced gene 20 protein) (SIG-20) |
| chr1_+_166428761 | 0.01 |
ENSRNOT00000085555
|
Stard10
|
StAR-related lipid transfer domain containing 10 |
| chr2_+_187977008 | 0.01 |
ENSRNOT00000081531
ENSRNOT00000092951 |
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr1_-_57327379 | 0.01 |
ENSRNOT00000080429
|
Dll1
|
delta like canonical Notch ligand 1 |
| chr10_-_88357025 | 0.01 |
ENSRNOT00000080006
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr7_-_139731876 | 0.01 |
ENSRNOT00000013704
|
Asb8
|
ankyrin repeat and SOCS box-containing 8 |
| chr7_+_123573930 | 0.00 |
ENSRNOT00000066149
|
Fam109b
|
family with sequence similarity 109, member B |
| chr9_-_81565416 | 0.00 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chr2_-_22059149 | 0.00 |
ENSRNOT00000031909
|
Fam151b
|
family with sequence similarity 151, member B |
| chr14_-_108412823 | 0.00 |
ENSRNOT00000081405
|
Pex13
|
peroxisomal biogenesis factor 13 |
| chr10_-_88356538 | 0.00 |
ENSRNOT00000022430
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr18_-_40716686 | 0.00 |
ENSRNOT00000000172
|
Cdo1
|
cysteine dioxygenase type 1 |
| chr6_+_145740035 | 0.00 |
ENSRNOT00000064868
|
Cdca7l
|
cell division cycle associated 7 like |
| chr20_-_12820466 | 0.00 |
ENSRNOT00000001699
|
Ftcd
|
formimidoyltransferase cyclodeaminase |
| chr10_-_49007089 | 0.00 |
ENSRNOT00000073706
|
Mapk7
|
mitogen-activated protein kinase 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0002489 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.0 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |