Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
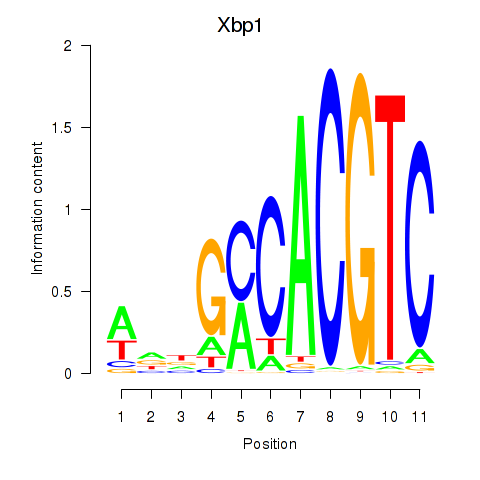
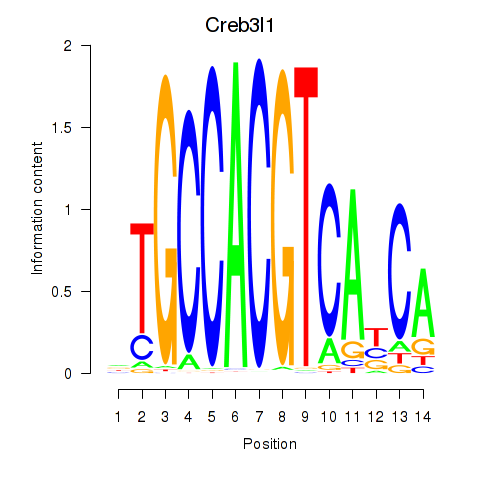
Results for Xbp1_Creb3l1
Z-value: 0.38
Transcription factors associated with Xbp1_Creb3l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Xbp1
|
ENSRNOG00000010298 | X-box binding protein 1 |
|
Creb3l1
|
ENSRNOG00000005413 | cAMP responsive element binding protein 3-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3l1 | rn6_v1_chr3_-_80933283_80933283 | 0.74 | 1.5e-01 | Click! |
| Xbp1 | rn6_v1_chr14_+_85753760_85753760 | -0.19 | 7.5e-01 | Click! |
Activity profile of Xbp1_Creb3l1 motif
Sorted Z-values of Xbp1_Creb3l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_41671437 | 0.28 |
ENSRNOT00000001883
|
Lhx5
|
LIM homeobox 5 |
| chr17_+_6909728 | 0.21 |
ENSRNOT00000061231
|
LOC681410
|
hypothetical protein LOC681410 |
| chr8_-_98738446 | 0.20 |
ENSRNOT00000019860
|
Zic1
|
Zic family member 1 |
| chr1_-_190965115 | 0.14 |
ENSRNOT00000023483
|
AC103221.1
|
|
| chr1_+_105284753 | 0.12 |
ENSRNOT00000041950
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr14_+_11198896 | 0.12 |
ENSRNOT00000079767
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr18_+_73157973 | 0.10 |
ENSRNOT00000031993
|
Skor2
|
SKI family transcriptional corepressor 2 |
| chr14_+_83752393 | 0.10 |
ENSRNOT00000081123
|
Selenom
|
selenoprotein M |
| chr3_+_14889510 | 0.09 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr1_+_199941161 | 0.09 |
ENSRNOT00000027616
|
Bag3
|
Bcl2-associated athanogene 3 |
| chr10_-_56558487 | 0.09 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr4_-_84768249 | 0.09 |
ENSRNOT00000013205
|
Fkbp14
|
FK506 binding protein 14 |
| chr10_-_105149971 | 0.08 |
ENSRNOT00000012436
|
Srp68
|
signal recognition particle 68 |
| chr1_+_221644867 | 0.08 |
ENSRNOT00000054830
|
Ehd1
|
EH-domain containing 1 |
| chr12_+_24473981 | 0.08 |
ENSRNOT00000001973
|
Fzd9
|
frizzled class receptor 9 |
| chr14_+_11199404 | 0.07 |
ENSRNOT00000003106
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr7_-_117978787 | 0.06 |
ENSRNOT00000074262
|
Zfp647
|
zinc finger protein 647 |
| chr11_+_64601029 | 0.06 |
ENSRNOT00000004138
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr18_-_47513030 | 0.06 |
ENSRNOT00000083881
ENSRNOT00000074226 |
Lox
|
lysyl oxidase |
| chr19_-_37938857 | 0.06 |
ENSRNOT00000026730
|
Slc12a4
|
solute carrier family 12 member 4 |
| chr18_+_52423883 | 0.06 |
ENSRNOT00000022017
|
Prrc1
|
proline-rich coiled-coil 1 |
| chr4_+_118852062 | 0.05 |
ENSRNOT00000090827
ENSRNOT00000025070 |
Gfpt1
|
glutamine fructose-6-phosphate transaminase 1 |
| chr4_-_82258765 | 0.05 |
ENSRNOT00000008523
|
Hoxa5
|
homeo box A5 |
| chr1_-_101086198 | 0.05 |
ENSRNOT00000027917
|
Rcn3
|
reticulocalbin 3 |
| chr4_+_56625561 | 0.05 |
ENSRNOT00000008356
ENSRNOT00000090808 ENSRNOT00000008972 |
Calu
|
calumenin |
| chr1_-_81144024 | 0.05 |
ENSRNOT00000088431
|
Zfp94
|
zinc finger protein 94 |
| chr16_+_20873642 | 0.04 |
ENSRNOT00000004891
|
Ddx49
|
DEAD-box helicase 49 |
| chr17_+_26925828 | 0.04 |
ENSRNOT00000018310
|
Txndc5
|
thioredoxin domain containing 5 |
| chr5_+_167952728 | 0.04 |
ENSRNOT00000085315
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr1_-_101085884 | 0.04 |
ENSRNOT00000085352
|
Rcn3
|
reticulocalbin 3 |
| chr3_-_10269693 | 0.04 |
ENSRNOT00000021871
|
Fubp3
|
far upstream element binding protein 3 |
| chr7_-_114305896 | 0.04 |
ENSRNOT00000067327
ENSRNOT00000038172 |
Trappc9
|
trafficking protein particle complex 9 |
| chr11_+_80255790 | 0.04 |
ENSRNOT00000002522
|
Bcl6
|
B-cell CLL/lymphoma 6 |
| chr15_-_4057104 | 0.04 |
ENSRNOT00000084350
ENSRNOT00000012270 |
Sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr9_-_82699551 | 0.04 |
ENSRNOT00000020673
|
Obsl1
|
obscurin-like 1 |
| chrX_+_156999826 | 0.03 |
ENSRNOT00000081819
|
Idh3g
|
isocitrate dehydrogenase 3 (NAD), gamma |
| chr4_+_342302 | 0.03 |
ENSRNOT00000009233
|
Insig1
|
insulin induced gene 1 |
| chr2_+_198823366 | 0.03 |
ENSRNOT00000083769
ENSRNOT00000028814 |
Pias3
|
protein inhibitor of activated STAT, 3 |
| chr11_-_64752544 | 0.03 |
ENSRNOT00000048738
|
Tmem39a
|
transmembrane protein 39a |
| chr5_+_59063531 | 0.03 |
ENSRNOT00000085311
ENSRNOT00000065909 |
Creb3
|
cAMP responsive element binding protein 3 |
| chr3_+_146582752 | 0.03 |
ENSRNOT00000010157
|
Pygb
|
glycogen phosphorylase B |
| chr2_-_183582553 | 0.03 |
ENSRNOT00000014236
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr17_+_27496353 | 0.03 |
ENSRNOT00000077994
ENSRNOT00000019040 ENSRNOT00000089736 |
Ssr1
|
signal sequence receptor subunit 1 |
| chr1_-_226255886 | 0.03 |
ENSRNOT00000027842
|
Fen1
|
flap structure-specific endonuclease 1 |
| chr11_+_46009322 | 0.03 |
ENSRNOT00000002233
|
Tmem45a
|
transmembrane protein 45A |
| chr9_-_100592271 | 0.03 |
ENSRNOT00000086433
|
Hdlbp
|
high density lipoprotein binding protein |
| chr4_+_55772377 | 0.03 |
ENSRNOT00000047698
|
Snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr3_-_151224123 | 0.03 |
ENSRNOT00000026091
|
Trpc4ap
|
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr10_-_84789832 | 0.03 |
ENSRNOT00000071719
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr3_+_160945359 | 0.03 |
ENSRNOT00000019761
|
Pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr1_+_123015746 | 0.03 |
ENSRNOT00000013483
|
Magel2
|
MAGE family member L2 |
| chr7_-_97759852 | 0.03 |
ENSRNOT00000007484
|
Derl1
|
derlin 1 |
| chr11_-_51202703 | 0.03 |
ENSRNOT00000002719
ENSRNOT00000077220 |
Cblb
|
Cbl proto-oncogene B |
| chr20_-_3822754 | 0.03 |
ENSRNOT00000000541
ENSRNOT00000077357 |
Slc39a7
|
solute carrier family 39 member 7 |
| chr3_-_138116664 | 0.03 |
ENSRNOT00000055602
|
Rrbp1
|
ribosome binding protein 1 |
| chr9_-_61810417 | 0.02 |
ENSRNOT00000020910
|
Rftn2
|
raftlin family member 2 |
| chr7_-_60742660 | 0.02 |
ENSRNOT00000086116
|
Mdm2
|
MDM2 proto-oncogene |
| chr7_+_117951154 | 0.02 |
ENSRNOT00000060181
|
Znf7
|
zinc finger protein 7 |
| chr12_-_13210758 | 0.02 |
ENSRNOT00000001438
|
Kdelr2
|
KDEL endoplasmic reticulum protein retention receptor 2 |
| chr17_-_1830001 | 0.02 |
ENSRNOT00000066230
|
Aaed1
|
AhpC/TSA antioxidant enzyme domain containing 1 |
| chr5_-_59063592 | 0.02 |
ENSRNOT00000022400
|
Tln1
|
talin 1 |
| chr16_-_20873344 | 0.02 |
ENSRNOT00000027381
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr18_-_24182012 | 0.02 |
ENSRNOT00000023594
|
Syt4
|
synaptotagmin 4 |
| chr3_+_11317183 | 0.02 |
ENSRNOT00000091171
ENSRNOT00000016341 |
Golga2
|
golgin A2 |
| chr1_-_216750844 | 0.02 |
ENSRNOT00000054862
|
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chrX_+_9815652 | 0.02 |
ENSRNOT00000091382
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr17_-_20364714 | 0.02 |
ENSRNOT00000070962
|
Jarid2
|
jumonji and AT-rich interaction domain containing 2 |
| chr10_-_105228547 | 0.02 |
ENSRNOT00000028875
|
Srp68
|
signal recognition particle 68 |
| chr2_+_227455722 | 0.02 |
ENSRNOT00000064809
|
Sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr9_+_100080144 | 0.01 |
ENSRNOT00000071411
|
Rnpepl1
|
arginyl aminopeptidase like 1 |
| chr7_+_29282305 | 0.01 |
ENSRNOT00000007623
|
Arl1
|
ADP-ribosylation factor like GTPase 1 |
| chr8_-_39243882 | 0.01 |
ENSRNOT00000082086
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr10_+_38877422 | 0.01 |
ENSRNOT00000065229
ENSRNOT00000083162 |
Sept8
|
septin 8 |
| chr18_+_1504178 | 0.01 |
ENSRNOT00000048198
|
Rpl36al
|
ribosomal protein L36a-like |
| chr7_-_11406771 | 0.01 |
ENSRNOT00000047450
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr5_-_59025631 | 0.01 |
ENSRNOT00000049000
ENSRNOT00000022801 |
Tpm2
|
tropomyosin 2, beta |
| chr1_-_87174530 | 0.01 |
ENSRNOT00000089578
|
Kcnk6
|
potassium two pore domain channel subfamily K member 6 |
| chr1_-_220067123 | 0.01 |
ENSRNOT00000071020
ENSRNOT00000072373 |
Rbm14
|
RNA binding motif protein 14 |
| chr8_-_21458379 | 0.01 |
ENSRNOT00000038084
|
Zfp26
|
zinc finger protein 26 |
| chr5_+_48274477 | 0.01 |
ENSRNOT00000075992
ENSRNOT00000041271 |
Ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr8_-_49075892 | 0.01 |
ENSRNOT00000082687
|
Arcn1
|
archain 1 |
| chr1_-_221448570 | 0.01 |
ENSRNOT00000075165
|
Zfpl1
|
zinc finger protein-like 1 |
| chr20_+_31298269 | 0.01 |
ENSRNOT00000000673
|
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr1_-_226152524 | 0.01 |
ENSRNOT00000027756
|
Fads2
|
fatty acid desaturase 2 |
| chr1_+_221448661 | 0.01 |
ENSRNOT00000072493
|
LOC100910252
|
sororin-like |
| chr2_+_124149872 | 0.01 |
ENSRNOT00000023584
|
Spata5
|
spermatogenesis associated 5 |
| chr6_+_38474804 | 0.01 |
ENSRNOT00000009779
|
Nbas
|
neuroblastoma amplified sequence |
| chr3_-_66394764 | 0.01 |
ENSRNOT00000075832
|
Cerkl
|
ceramide kinase-like |
| chr12_-_37480297 | 0.01 |
ENSRNOT00000001397
|
Tmed2
|
transmembrane p24 trafficking protein 2 |
| chr18_+_73226630 | 0.01 |
ENSRNOT00000051857
|
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr3_-_113091770 | 0.01 |
ENSRNOT00000068132
|
Lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr2_-_2891253 | 0.01 |
ENSRNOT00000029250
|
Arsk
|
arylsulfatase family, member K |
| chr4_+_119997268 | 0.01 |
ENSRNOT00000073429
|
Rpn1
|
ribophorin I |
| chr7_-_11444786 | 0.01 |
ENSRNOT00000027323
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr13_+_90467265 | 0.01 |
ENSRNOT00000008518
|
Copa
|
coatomer protein complex subunit alpha |
| chr17_+_30965942 | 0.01 |
ENSRNOT00000060143
|
Pxdc1
|
PX domain containing 1 |
| chr1_+_199360645 | 0.01 |
ENSRNOT00000026527
|
Kat8
|
lysine acetyltransferase 8 |
| chr1_+_80087684 | 0.01 |
ENSRNOT00000041215
|
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr3_-_79892429 | 0.01 |
ENSRNOT00000016191
|
Slc39a13
|
solute carrier family 39 member 13 |
| chr7_-_59547174 | 0.01 |
ENSRNOT00000085836
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr9_-_82673898 | 0.01 |
ENSRNOT00000027165
|
Chpf
|
chondroitin polymerizing factor |
| chr2_+_9526209 | 0.01 |
ENSRNOT00000021818
|
Lysmd3
|
LysM domain containing 3 |
| chr12_-_52558606 | 0.01 |
ENSRNOT00000056656
|
Golga3
|
golgin A3 |
| chr5_-_39650894 | 0.01 |
ENSRNOT00000010421
ENSRNOT00000089522 |
Ufl1
|
Ufm1-specific ligase 1 |
| chr1_-_87174821 | 0.01 |
ENSRNOT00000027974
|
Kcnk6
|
potassium two pore domain channel subfamily K member 6 |
| chrX_+_73390903 | 0.01 |
ENSRNOT00000077002
|
Zfp449
|
zinc finger protein 449 |
| chr6_+_134958854 | 0.01 |
ENSRNOT00000073950
ENSRNOT00000009377 |
Dync1h1
|
dynein cytoplasmic 1 heavy chain 1 |
| chr7_-_27136652 | 0.01 |
ENSRNOT00000086905
|
Hcfc2
|
host cell factor C2 |
| chr5_+_62109328 | 0.01 |
ENSRNOT00000012260
|
Nans
|
N-acetylneuraminate synthase |
| chr19_-_38120578 | 0.01 |
ENSRNOT00000026873
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr1_-_260517323 | 0.01 |
ENSRNOT00000018043
|
Tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr10_+_48569555 | 0.01 |
ENSRNOT00000003966
|
Adora2b
|
adenosine A2B receptor |
| chr8_+_112594691 | 0.01 |
ENSRNOT00000038383
ENSRNOT00000081281 |
Acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr3_+_59981959 | 0.01 |
ENSRNOT00000030460
|
Sp9
|
Sp9 transcription factor |
| chr1_+_225096598 | 0.01 |
ENSRNOT00000035650
|
Ganab
|
glucosidase II alpha subunit |
| chr13_-_90467235 | 0.01 |
ENSRNOT00000081534
ENSRNOT00000008150 |
Ncstn
|
nicastrin |
| chr1_-_78851719 | 0.00 |
ENSRNOT00000022603
|
Calm2
|
calmodulin 2 |
| chr4_-_120467932 | 0.00 |
ENSRNOT00000018454
|
Sec61a1
|
Sec61 translocon alpha 1 subunit |
| chr8_-_22111863 | 0.00 |
ENSRNOT00000046983
|
Raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr20_+_3823042 | 0.00 |
ENSRNOT00000041613
|
Rxrb
|
retinoid X receptor beta |
| chr19_-_49448072 | 0.00 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr9_+_82674202 | 0.00 |
ENSRNOT00000027208
|
Tmem198
|
transmembrane protein 198 |
| chr4_-_145699577 | 0.00 |
ENSRNOT00000014377
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr6_+_91476698 | 0.00 |
ENSRNOT00000005608
|
Mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr14_-_34528262 | 0.00 |
ENSRNOT00000003010
|
Tmem165
|
transmembrane protein 165 |
| chr7_+_31784438 | 0.00 |
ENSRNOT00000010914
ENSRNOT00000010929 |
Ikbip
|
IKBKB interacting protein |
| chr3_+_95232166 | 0.00 |
ENSRNOT00000017952
|
LOC691083
|
hypothetical protein LOC691083 |
| chr11_+_46714355 | 0.00 |
ENSRNOT00000073404
|
Tmem45al
|
transmembrane protein 45A-like |
| chr8_-_13109487 | 0.00 |
ENSRNOT00000061616
|
Amotl1
|
angiomotin-like 1 |
| chrX_-_156999650 | 0.00 |
ENSRNOT00000083557
|
Ssr4
|
signal sequence receptor subunit 4 |
| chr20_+_46250363 | 0.00 |
ENSRNOT00000076522
ENSRNOT00000000334 |
Cd164
|
CD164 molecule |
| chr7_+_24939498 | 0.00 |
ENSRNOT00000010601
|
Ckap4
|
cytoskeleton-associated protein 4 |
| chr15_-_57805184 | 0.00 |
ENSRNOT00000000168
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr12_+_47218969 | 0.00 |
ENSRNOT00000081343
ENSRNOT00000038395 |
Unc119b
|
unc-119 lipid binding chaperone B |
| chr18_+_27114822 | 0.00 |
ENSRNOT00000027418
|
Srp19
|
signal recognition particle 19 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Xbp1_Creb3l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) |
| 0.0 | 0.1 | GO:1901073 | glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.1 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.0 | 0.0 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.0 | GO:1903243 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.0 | GO:0045242 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |