Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Vsx2_Dlx3
Z-value: 0.18
Transcription factors associated with Vsx2_Dlx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
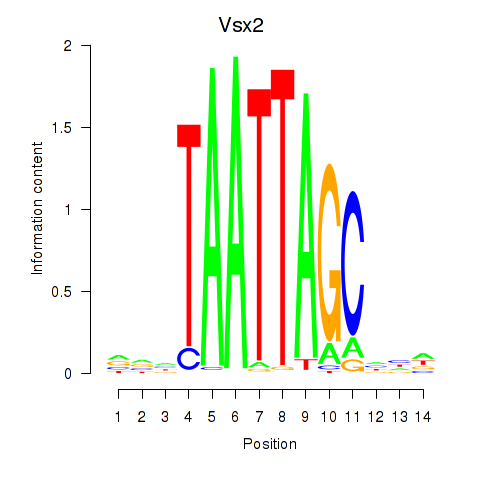
Vsx2
|
ENSRNOG00000011918 | visual system homeobox 2 |
|
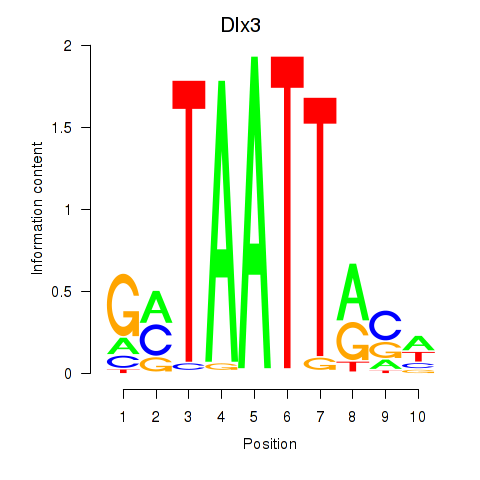
Dlx3
|
ENSRNOG00000004278 | distal-less homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Vsx2 | rn6_v1_chr6_+_108285822_108285822 | 0.71 | 1.8e-01 | Click! |
| Dlx3 | rn6_v1_chr10_+_82937971_82937971 | -0.60 | 2.8e-01 | Click! |
Activity profile of Vsx2_Dlx3 motif
Sorted Z-values of Vsx2_Dlx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_17186679 | 0.42 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr12_-_2174131 | 0.37 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr3_-_66417741 | 0.14 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr2_+_145174876 | 0.10 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr18_-_26211445 | 0.09 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr4_+_94696965 | 0.09 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr10_-_18131562 | 0.08 |
ENSRNOT00000006676
|
Tlx3
|
T-cell leukemia, homeobox 3 |
| chr10_+_103395511 | 0.08 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr13_+_31081804 | 0.04 |
ENSRNOT00000041413
|
Cdh7
|
cadherin 7 |
| chr13_+_71107465 | 0.04 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr6_+_58468155 | 0.03 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr3_+_95715193 | 0.03 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr1_-_124803363 | 0.02 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr4_-_117575154 | 0.02 |
ENSRNOT00000075813
|
LOC102556148
|
probable N-acetyltransferase CML2-like |
| chr12_-_35979193 | 0.02 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr12_-_45801842 | 0.02 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr10_+_91217079 | 0.02 |
ENSRNOT00000004218
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr12_-_19167015 | 0.02 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chr20_+_44680449 | 0.02 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr17_-_2705123 | 0.02 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr6_-_111417813 | 0.02 |
ENSRNOT00000016324
ENSRNOT00000085458 |
Sptlc2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr7_-_142348232 | 0.02 |
ENSRNOT00000078848
|
Galnt6
|
polypeptide N-acetylgalactosaminyltransferase 6 |
| chr18_-_26656879 | 0.01 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr5_-_145423172 | 0.01 |
ENSRNOT00000081919
|
Gjb5
|
gap junction protein, beta 5 |
| chr1_+_217345545 | 0.01 |
ENSRNOT00000071741
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr5_-_139748489 | 0.01 |
ENSRNOT00000078741
|
Nfyc
|
nuclear transcription factor Y subunit gamma |
| chr12_-_10335499 | 0.01 |
ENSRNOT00000071567
|
Wasf3
|
WAS protein family, member 3 |
| chr4_-_17594598 | 0.01 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr2_-_30634243 | 0.01 |
ENSRNOT00000077537
|
Marveld2
|
MARVEL domain containing 2 |
| chr16_+_35934970 | 0.01 |
ENSRNOT00000084707
ENSRNOT00000016474 |
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr7_-_2941122 | 0.01 |
ENSRNOT00000082107
|
Esyt1
|
extended synaptotagmin 1 |
| chr4_-_4473307 | 0.01 |
ENSRNOT00000045773
|
Dpp6
|
dipeptidyl peptidase like 6 |
| chr1_+_13595295 | 0.01 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr1_+_193076430 | 0.01 |
ENSRNOT00000086492
ENSRNOT00000017885 |
Tnrc6a
|
trinucleotide repeat containing 6a |
| chr6_-_115616766 | 0.01 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chr8_+_81863619 | 0.01 |
ENSRNOT00000080608
|
Fam214a
|
family with sequence similarity 214, member A |
| chr10_-_87261717 | 0.01 |
ENSRNOT00000015740
|
Krt27
|
keratin 27 |
| chr14_+_76732650 | 0.01 |
ENSRNOT00000088197
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr3_+_47578384 | 0.01 |
ENSRNOT00000082769
|
Psmd14
|
proteasome 26S subunit, non-ATPase 14 |
| chr8_+_47674321 | 0.01 |
ENSRNOT00000026170
ENSRNOT00000078776 |
Trim29
|
tripartite motif-containing 29 |
| chr5_-_139749050 | 0.01 |
ENSRNOT00000056651
|
Nfyc
|
nuclear transcription factor Y subunit gamma |
| chr18_+_51523758 | 0.01 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr12_+_18679789 | 0.01 |
ENSRNOT00000001863
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr13_-_100672731 | 0.01 |
ENSRNOT00000004319
|
Degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr4_-_21920651 | 0.01 |
ENSRNOT00000066211
|
Tmem243
|
transmembrane protein 243 |
| chr20_-_27578244 | 0.01 |
ENSRNOT00000000708
|
Fam26d
|
family with sequence similarity 26, member D |
| chr9_-_53315915 | 0.01 |
ENSRNOT00000038093
|
Mstn
|
myostatin |
| chr3_+_117421604 | 0.01 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr20_+_29655226 | 0.01 |
ENSRNOT00000089059
|
Spock2
|
SPARC/osteonectin, cwcv and kazal like domains proteoglycan 2 |
| chr2_+_226563050 | 0.01 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr1_+_99599620 | 0.01 |
ENSRNOT00000024690
|
Ctu1
|
cytosolic thiouridylase subunit 1 |
| chr8_+_48718329 | 0.01 |
ENSRNOT00000089763
|
Slc37a4
|
solute carrier family 37 member 4 |
| chr2_+_80269661 | 0.01 |
ENSRNOT00000015975
|
AABR07008940.1
|
|
| chr7_-_107391184 | 0.01 |
ENSRNOT00000056793
|
Tmem71
|
transmembrane protein 71 |
| chr2_-_33025271 | 0.01 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr4_-_150485781 | 0.01 |
ENSRNOT00000008763
|
Zfp248
|
zinc finger protein 248 |
| chr2_+_187951344 | 0.01 |
ENSRNOT00000027123
|
Ssr2
|
signal sequence receptor, beta |
| chr9_-_38495126 | 0.01 |
ENSRNOT00000016933
ENSRNOT00000090385 |
Rab23
|
RAB23, member RAS oncogene family |
| chr6_-_123577695 | 0.01 |
ENSRNOT00000006604
|
Foxn3
|
forkhead box N3 |
| chr3_+_95233874 | 0.01 |
ENSRNOT00000079743
|
LOC691083
|
hypothetical protein LOC691083 |
| chr20_-_3728844 | 0.00 |
ENSRNOT00000074958
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 |
| chr18_+_79773608 | 0.00 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr7_+_3173287 | 0.00 |
ENSRNOT00000092037
|
Pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr5_-_150704117 | 0.00 |
ENSRNOT00000067455
|
Sesn2
|
sestrin 2 |
| chr1_-_220938814 | 0.00 |
ENSRNOT00000028081
|
Ovol1
|
ovo like transcriptional repressor 1 |
| chr12_+_46042413 | 0.00 |
ENSRNOT00000046882
|
Ccdc60
|
coiled-coil domain containing 60 |
| chrX_+_136466779 | 0.00 |
ENSRNOT00000093268
ENSRNOT00000068717 |
Arhgap36
|
Rho GTPase activating protein 36 |
| chr18_+_16544508 | 0.00 |
ENSRNOT00000020601
|
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr4_-_100252755 | 0.00 |
ENSRNOT00000017301
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr4_-_50312608 | 0.00 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr9_+_71915421 | 0.00 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr5_-_28131133 | 0.00 |
ENSRNOT00000067331
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr20_-_25826658 | 0.00 |
ENSRNOT00000057950
ENSRNOT00000084660 |
Ctnna3
|
catenin alpha 3 |
| chr7_+_2752680 | 0.00 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr4_+_31333970 | 0.00 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr1_+_127802978 | 0.00 |
ENSRNOT00000055877
|
Adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr13_+_51229890 | 0.00 |
ENSRNOT00000060669
|
Cyb5r1
|
cytochrome b5 reductase 1 |
| chr8_+_36766977 | 0.00 |
ENSRNOT00000017648
|
Pus3
|
pseudouridylate synthase 3 |
| chr1_-_101095594 | 0.00 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr13_+_90943255 | 0.00 |
ENSRNOT00000011539
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr7_+_38900593 | 0.00 |
ENSRNOT00000072166
|
Epyc
|
epiphycan |
| chr2_-_113616766 | 0.00 |
ENSRNOT00000016858
ENSRNOT00000074723 |
Tmem212
|
transmembrane protein 212 |
| chr8_+_103938520 | 0.00 |
ENSRNOT00000067835
|
Gk5
|
glycerol kinase 5 (putative) |
| chr7_+_78558701 | 0.00 |
ENSRNOT00000006393
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr1_-_131460473 | 0.00 |
ENSRNOT00000084336
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr19_-_17399548 | 0.00 |
ENSRNOT00000075991
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr9_+_81566074 | 0.00 |
ENSRNOT00000074131
ENSRNOT00000046229 ENSRNOT00000090383 |
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr2_-_158133861 | 0.00 |
ENSRNOT00000090700
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr16_-_73152921 | 0.00 |
ENSRNOT00000048602
|
Zmat4
|
zinc finger, matrin type 4 |
| chr15_-_27819376 | 0.00 |
ENSRNOT00000067400
|
A930018M24Rik
|
RIKEN cDNA A930018M24 gene |
| chr7_-_122581777 | 0.00 |
ENSRNOT00000025758
|
Slc25a17
|
solute carrier family 25 member 17 |
| chr14_-_19159923 | 0.00 |
ENSRNOT00000003879
|
Afp
|
alpha-fetoprotein |
| chr3_+_48106099 | 0.00 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr10_+_82032656 | 0.00 |
ENSRNOT00000067751
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr14_+_39964588 | 0.00 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr17_-_45746753 | 0.00 |
ENSRNOT00000077475
|
RGD1564243
|
similar to brain Zn-finger protein |
| chr13_+_76962504 | 0.00 |
ENSRNOT00000076581
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr2_+_266315036 | 0.00 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr13_+_98311827 | 0.00 |
ENSRNOT00000082844
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr11_-_46693565 | 0.00 |
ENSRNOT00000044900
|
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr7_-_83348487 | 0.00 |
ENSRNOT00000006685
|
Nudcd1
|
NudC domain containing 1 |
| chr9_-_81566096 | 0.00 |
ENSRNOT00000019497
|
Aamp
|
angio-associated, migratory cell protein |
| chrX_+_124321551 | 0.00 |
ENSRNOT00000074486
|
LOC100910807
|
transcriptional regulator Kaiso-like |
| chr14_-_21128505 | 0.00 |
ENSRNOT00000004776
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr7_+_83348694 | 0.00 |
ENSRNOT00000006402
|
Eny2
|
ENY2, transcription and export complex 2 subunit |
| chr4_-_28437676 | 0.00 |
ENSRNOT00000012995
|
Hepacam2
|
HEPACAM family member 2 |
| chr1_+_185210922 | 0.00 |
ENSRNOT00000055120
|
Pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr1_+_98414226 | 0.00 |
ENSRNOT00000090785
|
Siglecl1
|
SIGLEC family like 1 |
| chr20_-_2224463 | 0.00 |
ENSRNOT00000001019
|
Trim26
|
tripartite motif-containing 26 |
| chr9_-_65736852 | 0.00 |
ENSRNOT00000087458
|
Trak2
|
trafficking kinesin protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Vsx2_Dlx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.0 | GO:0021917 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |