Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
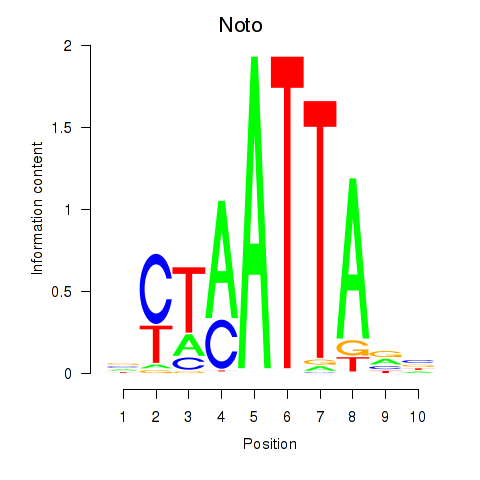
Results for Vsx1_Uncx_Prrx2_Shox2_Noto
Z-value: 0.98
Transcription factors associated with Vsx1_Uncx_Prrx2_Shox2_Noto
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
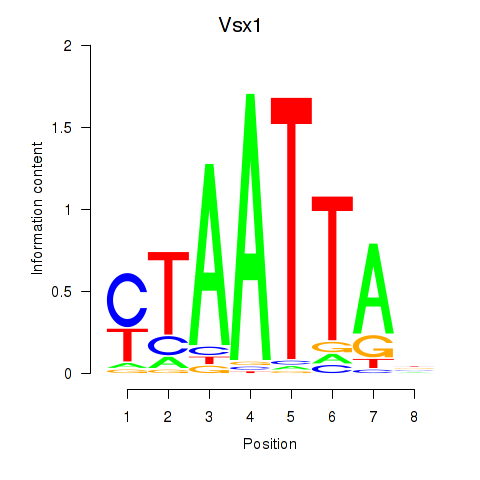
Vsx1
|
ENSRNOG00000042220 | visual system homeobox 1 |
|
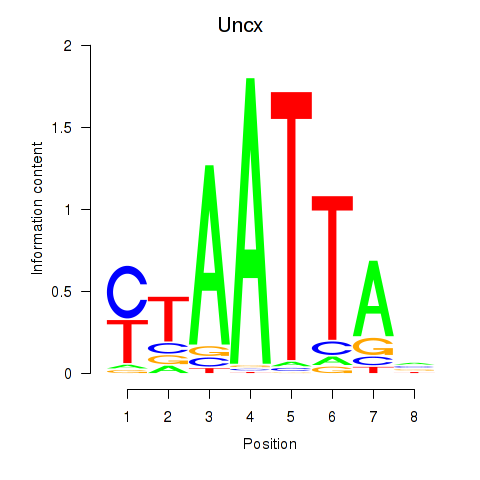
Uncx
|
ENSRNOG00000001284 | UNC homeobox |
|
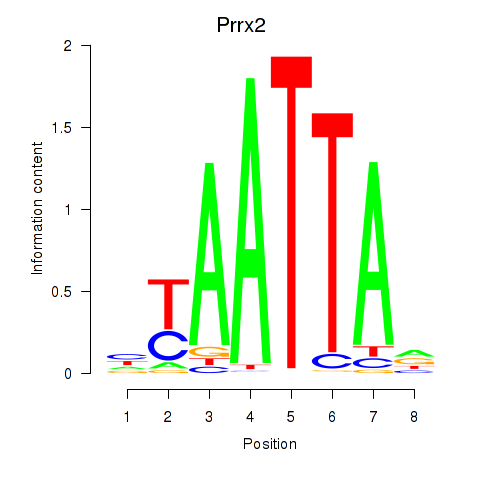
Prrx2
|
ENSRNOG00000058329 | paired related homeobox 2 |
|
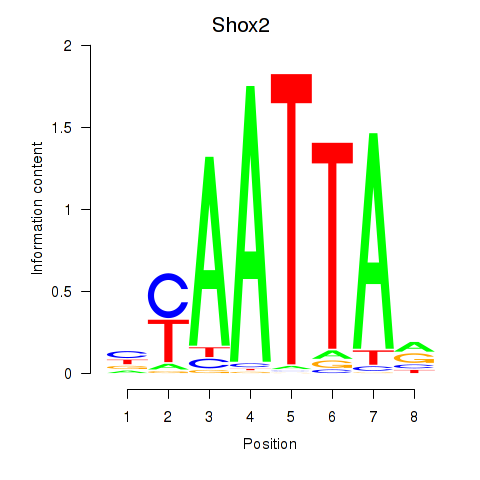
Shox2
|
ENSRNOG00000012478 | short stature homeobox 2 |
|
Noto
|
ENSRNOG00000037863 | notochord homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Uncx | rn6_v1_chr12_-_17186679_17186679 | 0.76 | 1.4e-01 | Click! |
| Shox2 | rn6_v1_chr2_-_164126783_164126783 | 0.65 | 2.3e-01 | Click! |
| Prrx2 | rn6_v1_chr3_+_9681871_9681871 | 0.28 | 6.5e-01 | Click! |
| Vsx1 | rn6_v1_chr3_-_146491837_146491837 | -0.08 | 9.0e-01 | Click! |
Activity profile of Vsx1_Uncx_Prrx2_Shox2_Noto motif
Sorted Z-values of Vsx1_Uncx_Prrx2_Shox2_Noto motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_2174131 | 6.90 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr10_-_88670430 | 2.25 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr2_+_145174876 | 1.66 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr12_-_17186679 | 1.35 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr4_-_80395502 | 1.10 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chr2_-_185852759 | 0.71 |
ENSRNOT00000049461
|
Mab21l2
|
mab-21 like 2 |
| chr17_+_11683862 | 0.68 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr1_+_105284753 | 0.60 |
ENSRNOT00000041950
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr10_-_87232723 | 0.49 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr6_-_125723732 | 0.49 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr3_+_95715193 | 0.44 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr4_-_180234804 | 0.40 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr3_-_7498555 | 0.38 |
ENSRNOT00000017725
|
Barhl1
|
BarH-like homeobox 1 |
| chr5_+_50381244 | 0.33 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr6_-_125723944 | 0.32 |
ENSRNOT00000007004
|
Fbln5
|
fibulin 5 |
| chr8_+_71914867 | 0.31 |
ENSRNOT00000023372
|
Dapk2
|
death-associated protein kinase 2 |
| chr14_+_4362717 | 0.29 |
ENSRNOT00000002887
|
Barhl2
|
BarH-like homeobox 2 |
| chr1_-_173764246 | 0.29 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chr2_+_196334626 | 0.27 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr6_+_28382962 | 0.27 |
ENSRNOT00000016976
|
Pomc
|
proopiomelanocortin |
| chr10_-_87286387 | 0.25 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr10_+_103395511 | 0.23 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr3_+_14889510 | 0.23 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr3_+_5709236 | 0.22 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr6_-_106971250 | 0.22 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr12_-_45801842 | 0.21 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr13_+_49005405 | 0.18 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr7_-_28711761 | 0.17 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr7_+_136182224 | 0.17 |
ENSRNOT00000008159
|
Tmem117
|
transmembrane protein 117 |
| chr1_-_124803363 | 0.16 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr3_+_45683993 | 0.15 |
ENSRNOT00000038983
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr20_+_44680449 | 0.15 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr3_-_57607683 | 0.15 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chr2_+_239415046 | 0.13 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr1_-_48825364 | 0.13 |
ENSRNOT00000024213
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr15_-_95514259 | 0.13 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr4_+_22898527 | 0.13 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr5_-_12526962 | 0.13 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr14_+_42714315 | 0.13 |
ENSRNOT00000084095
ENSRNOT00000091449 |
Phox2b
|
paired-like homeobox 2b |
| chr2_-_45518502 | 0.12 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr3_-_141411170 | 0.12 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr17_-_14627937 | 0.12 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chr16_+_54358471 | 0.12 |
ENSRNOT00000047314
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chrX_-_124464963 | 0.11 |
ENSRNOT00000036472
ENSRNOT00000077697 |
Tmem255a
|
transmembrane protein 255A |
| chr1_+_101178104 | 0.11 |
ENSRNOT00000028072
|
Pth2
|
parathyroid hormone 2 |
| chr4_-_100252755 | 0.11 |
ENSRNOT00000017301
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr9_-_85243001 | 0.10 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr6_-_7058314 | 0.10 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr6_-_67084234 | 0.10 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr7_+_72924799 | 0.09 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr6_+_58468155 | 0.09 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr13_+_71107465 | 0.08 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chrX_-_118998300 | 0.08 |
ENSRNOT00000025575
|
AABR07041089.1
|
|
| chr10_+_91217079 | 0.08 |
ENSRNOT00000004218
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr2_+_80269661 | 0.08 |
ENSRNOT00000015975
|
AABR07008940.1
|
|
| chr4_+_158088505 | 0.08 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr1_+_101599018 | 0.08 |
ENSRNOT00000028494
|
Fut1
|
fucosyltransferase 1 |
| chr7_-_107616038 | 0.08 |
ENSRNOT00000088752
|
Sla
|
src-like adaptor |
| chr6_-_111417813 | 0.07 |
ENSRNOT00000016324
ENSRNOT00000085458 |
Sptlc2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr3_-_161917285 | 0.07 |
ENSRNOT00000025150
|
Cdh22
|
cadherin 22 |
| chrX_+_118197217 | 0.07 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr3_-_11452529 | 0.07 |
ENSRNOT00000020206
|
Slc25a25
|
solute carrier family 25 member 25 |
| chr2_-_123972356 | 0.07 |
ENSRNOT00000023348
|
Il21
|
interleukin 21 |
| chrX_+_6273733 | 0.07 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr4_+_180062799 | 0.07 |
ENSRNOT00000021623
|
Rassf8
|
Ras association domain family member 8 |
| chr13_-_32427177 | 0.07 |
ENSRNOT00000044628
|
Cdh19
|
cadherin 19 |
| chr2_+_158097843 | 0.07 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr2_+_51672722 | 0.06 |
ENSRNOT00000016485
|
Fgf10
|
fibroblast growth factor 10 |
| chr7_+_3173287 | 0.06 |
ENSRNOT00000092037
|
Pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr10_-_84920886 | 0.06 |
ENSRNOT00000068083
|
Sp2
|
Sp2 transcription factor |
| chr1_-_215033460 | 0.06 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr9_-_18371856 | 0.06 |
ENSRNOT00000027293
|
Supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr5_-_150704117 | 0.06 |
ENSRNOT00000067455
|
Sesn2
|
sestrin 2 |
| chr9_-_82146874 | 0.06 |
ENSRNOT00000024190
|
Fev
|
FEV, ETS transcription factor |
| chr1_-_90149991 | 0.06 |
ENSRNOT00000076987
|
RGD1308428
|
similar to RIKEN cDNA 4931406P16 |
| chr4_-_17594598 | 0.06 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr4_-_18396035 | 0.06 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr9_+_10941613 | 0.06 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr19_+_53044379 | 0.06 |
ENSRNOT00000072369
|
Foxc2
|
forkhead box C2 |
| chr2_+_210880777 | 0.06 |
ENSRNOT00000026237
|
Gnat2
|
G protein subunit alpha transducin 2 |
| chr4_+_68656928 | 0.06 |
ENSRNOT00000016232
|
Tas2r108
|
taste receptor, type 2, member 108 |
| chr2_-_32518643 | 0.06 |
ENSRNOT00000061032
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_198755262 | 0.06 |
ENSRNOT00000028807
|
Rbm8a
|
RNA binding motif protein 8A |
| chr16_+_7035241 | 0.06 |
ENSRNOT00000084977
|
Nek4
|
NIMA-related kinase 4 |
| chr9_+_73418607 | 0.05 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr15_-_37830131 | 0.05 |
ENSRNOT00000075861
|
Eef1akmt1
|
eukaryotic translation elongation factor 1 alpha lysine methyltransferase 1 |
| chr10_+_17579284 | 0.05 |
ENSRNOT00000079698
|
Fbxw11
|
F-box and WD repeat domain containing 11 |
| chr3_-_44086006 | 0.05 |
ENSRNOT00000034449
ENSRNOT00000082604 |
Ermn
|
ermin |
| chr15_+_108318664 | 0.05 |
ENSRNOT00000057469
|
Ubac2
|
UBA domain containing 2 |
| chr2_-_58534211 | 0.05 |
ENSRNOT00000089178
|
Skp2
|
S-phase kinase associated protein 2 |
| chr18_-_49937522 | 0.05 |
ENSRNOT00000033254
|
Zfp608
|
zinc finger protein 608 |
| chr16_+_63958645 | 0.05 |
ENSRNOT00000081522
ENSRNOT00000082355 ENSRNOT00000013991 |
Nrg1
|
neuregulin 1 |
| chr8_+_47674321 | 0.05 |
ENSRNOT00000026170
ENSRNOT00000078776 |
Trim29
|
tripartite motif-containing 29 |
| chr14_-_16903242 | 0.05 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr9_+_81566074 | 0.05 |
ENSRNOT00000074131
ENSRNOT00000046229 ENSRNOT00000090383 |
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chrX_+_33443186 | 0.04 |
ENSRNOT00000005622
|
S100g
|
S100 calcium binding protein G |
| chr18_-_26656879 | 0.04 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chrX_-_104932508 | 0.04 |
ENSRNOT00000075325
|
Nox1
|
NADPH oxidase 1 |
| chrX_+_156463953 | 0.04 |
ENSRNOT00000079889
|
Flna
|
filamin A |
| chr5_-_136965191 | 0.04 |
ENSRNOT00000056842
|
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr10_-_88172910 | 0.04 |
ENSRNOT00000046956
|
Krt42
|
keratin 42 |
| chr1_+_13595295 | 0.04 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr1_-_15374850 | 0.04 |
ENSRNOT00000016728
|
Pex7
|
peroxisomal biogenesis factor 7 |
| chr1_+_185210922 | 0.04 |
ENSRNOT00000055120
|
Pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr7_-_101138373 | 0.04 |
ENSRNOT00000043257
|
LOC500877
|
Ab1-152 |
| chr1_-_134871167 | 0.04 |
ENSRNOT00000076300
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chrX_-_80142902 | 0.04 |
ENSRNOT00000071650
|
Hmgn5
|
high mobility group nucleosome binding domain 5 |
| chr15_+_11298478 | 0.04 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr15_+_44441856 | 0.04 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr14_+_76732650 | 0.04 |
ENSRNOT00000088197
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr5_+_139790395 | 0.03 |
ENSRNOT00000015033
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr9_-_53315915 | 0.03 |
ENSRNOT00000038093
|
Mstn
|
myostatin |
| chr2_+_226563050 | 0.03 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr2_+_154580558 | 0.03 |
ENSRNOT00000076976
|
Gmps
|
guanine monophosphate synthase |
| chr8_-_53816447 | 0.03 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr5_-_28131133 | 0.03 |
ENSRNOT00000067331
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr13_-_99101208 | 0.03 |
ENSRNOT00000004329
|
H3f3a
|
H3 histone family member 3A |
| chr18_+_867048 | 0.03 |
ENSRNOT00000065494
|
Colec12
|
collectin sub-family member 12 |
| chr5_+_129756149 | 0.03 |
ENSRNOT00000067277
|
Dmrta2
|
DMRT-like family A2 |
| chr9_+_88493593 | 0.03 |
ENSRNOT00000020712
|
LOC102556337
|
mitochondrial fission factor-like |
| chr19_+_39229754 | 0.03 |
ENSRNOT00000050612
|
Vps4a
|
vacuolar protein sorting 4 homolog A |
| chrX_+_159158194 | 0.03 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr19_-_49448072 | 0.03 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr18_+_51523758 | 0.03 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr2_-_235249571 | 0.03 |
ENSRNOT00000093631
ENSRNOT00000085096 |
Rrh
|
retinal pigment epithelium derived rhodopsin homolog |
| chr2_+_225645568 | 0.03 |
ENSRNOT00000017878
|
Abca4
|
ATP binding cassette subfamily A member 4 |
| chr4_-_56786754 | 0.03 |
ENSRNOT00000050795
|
Kcp
|
kielin/chordin-like protein |
| chr18_-_61057365 | 0.03 |
ENSRNOT00000042352
|
Alpk2
|
alpha-kinase 2 |
| chr4_-_4473307 | 0.03 |
ENSRNOT00000045773
|
Dpp6
|
dipeptidyl peptidase like 6 |
| chr5_+_58855773 | 0.03 |
ENSRNOT00000072869
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr10_-_93675991 | 0.03 |
ENSRNOT00000090662
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr11_-_4397361 | 0.03 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr18_-_12640716 | 0.03 |
ENSRNOT00000020697
|
Klhl14
|
kelch-like family member 14 |
| chr4_-_157304653 | 0.03 |
ENSRNOT00000051613
|
Lrrc23
|
leucine rich repeat containing 23 |
| chr1_+_264741911 | 0.03 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr7_-_142062870 | 0.02 |
ENSRNOT00000026531
|
Slc11a2
|
solute carrier family 11 member 2 |
| chr1_+_193076430 | 0.02 |
ENSRNOT00000086492
ENSRNOT00000017885 |
Tnrc6a
|
trinucleotide repeat containing 6a |
| chr3_+_113319456 | 0.02 |
ENSRNOT00000051354
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr11_-_32550539 | 0.02 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chr20_-_3728844 | 0.02 |
ENSRNOT00000074958
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 |
| chr17_-_77527894 | 0.02 |
ENSRNOT00000032173
|
Bend7
|
BEN domain containing 7 |
| chr15_+_31950986 | 0.02 |
ENSRNOT00000080233
|
AABR07017868.4
|
|
| chr2_-_88553086 | 0.02 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr2_+_102685513 | 0.02 |
ENSRNOT00000033940
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr3_-_102826379 | 0.02 |
ENSRNOT00000073423
|
Olr769
|
olfactory receptor 769 |
| chr13_+_87986240 | 0.02 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr8_+_103938520 | 0.02 |
ENSRNOT00000067835
|
Gk5
|
glycerol kinase 5 (putative) |
| chr16_-_74264142 | 0.02 |
ENSRNOT00000026067
|
Dkk4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr20_+_45458558 | 0.02 |
ENSRNOT00000000713
|
Cdk19
|
cyclin-dependent kinase 19 |
| chr3_-_37803112 | 0.02 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr6_+_146784915 | 0.02 |
ENSRNOT00000008362
|
Sp8
|
Sp8 transcription factor |
| chr12_-_35979193 | 0.02 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr4_-_21920651 | 0.02 |
ENSRNOT00000066211
|
Tmem243
|
transmembrane protein 243 |
| chr7_+_44009069 | 0.02 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr14_-_44078897 | 0.02 |
ENSRNOT00000031792
|
N4bp2
|
NEDD4 binding protein 2 |
| chr11_+_13499164 | 0.02 |
ENSRNOT00000013159
|
LOC680121
|
similar to heat shock protein 8 |
| chr16_+_4469468 | 0.02 |
ENSRNOT00000021164
|
Wnt5a
|
wingless-type MMTV integration site family, member 5A |
| chr1_+_277355619 | 0.02 |
ENSRNOT00000022788
|
Nhlrc2
|
NHL repeat containing 2 |
| chr1_+_127802978 | 0.02 |
ENSRNOT00000055877
|
Adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr10_-_91986632 | 0.02 |
ENSRNOT00000087824
|
Nsf
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr7_+_42269784 | 0.02 |
ENSRNOT00000008471
ENSRNOT00000007231 |
Kitlg
|
KIT ligand |
| chr9_-_81566096 | 0.02 |
ENSRNOT00000019497
|
Aamp
|
angio-associated, migratory cell protein |
| chr1_-_102699442 | 0.02 |
ENSRNOT00000056109
|
Tph1
|
tryptophan hydroxylase 1 |
| chr9_+_20241062 | 0.01 |
ENSRNOT00000071593
|
LOC100911585
|
leucine-rich repeat-containing protein 23-like |
| chr6_+_2216623 | 0.01 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr6_-_115616766 | 0.01 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chr11_-_76888178 | 0.01 |
ENSRNOT00000049841
|
Ostn
|
osteocrin |
| chr3_+_2591331 | 0.01 |
ENSRNOT00000017466
|
Sapcd2
|
suppressor APC domain containing 2 |
| chr3_+_95233874 | 0.01 |
ENSRNOT00000079743
|
LOC691083
|
hypothetical protein LOC691083 |
| chr10_-_87195075 | 0.01 |
ENSRNOT00000014851
|
Krt24
|
keratin 24 |
| chr9_+_79630604 | 0.01 |
ENSRNOT00000021539
ENSRNOT00000083979 |
Tmem169
|
transmembrane protein 169 |
| chr18_-_32670665 | 0.01 |
ENSRNOT00000019409
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr1_-_134867001 | 0.01 |
ENSRNOT00000090149
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr16_+_50049828 | 0.01 |
ENSRNOT00000034448
|
Fam149a
|
family with sequence similarity 149, member A |
| chr7_+_2752680 | 0.01 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr9_-_79630452 | 0.01 |
ENSRNOT00000078125
ENSRNOT00000086044 ENSRNOT00000089283 |
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr6_+_125876109 | 0.01 |
ENSRNOT00000091300
|
Cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr12_+_39423596 | 0.01 |
ENSRNOT00000066952
|
Ift81
|
intraflagellar transport 81 |
| chr3_-_45169118 | 0.01 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr6_-_23316962 | 0.01 |
ENSRNOT00000065421
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr11_+_45751812 | 0.01 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr3_+_10036461 | 0.01 |
ENSRNOT00000073731
|
Qrfp
|
pyroglutamylated RFamide peptide |
| chr7_-_15073052 | 0.01 |
ENSRNOT00000037708
|
Zfp799
|
zinc finger protein 799 |
| chr3_+_110855000 | 0.01 |
ENSRNOT00000081613
|
Knl1
|
kinetochore scaffold 1 |
| chr5_-_28130803 | 0.01 |
ENSRNOT00000093186
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr10_+_54352270 | 0.01 |
ENSRNOT00000036752
|
Dhrs7c
|
dehydrogenase/reductase 7C |
| chr10_+_53740841 | 0.01 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr3_+_138508899 | 0.01 |
ENSRNOT00000009437
|
Kat14
|
lysine acetyltransferase 14 |
| chr11_+_80742467 | 0.01 |
ENSRNOT00000002507
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr8_+_23193181 | 0.01 |
ENSRNOT00000071703
|
Zfp872
|
zinc finger protein 872 |
| chr3_+_9403840 | 0.01 |
ENSRNOT00000071380
|
NEWGENE_735020
|
pyroglutamylated RFamide peptide |
| chr4_+_61924013 | 0.01 |
ENSRNOT00000090717
|
Bpgm
|
bisphosphoglycerate mutase |
| chr15_+_4064706 | 0.01 |
ENSRNOT00000011956
|
Synpo2l
|
synaptopodin 2-like |
| chr7_+_42304534 | 0.00 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr19_+_38039729 | 0.00 |
ENSRNOT00000089783
|
Nfatc3
|
nuclear factor of activated T-cells 3 |
| chr2_+_118910587 | 0.00 |
ENSRNOT00000065518
|
Zfp639
|
zinc finger protein 639 |
| chr2_-_248484100 | 0.00 |
ENSRNOT00000040856
|
Gbp3
|
guanylate binding protein 3 |
| chr4_-_169036950 | 0.00 |
ENSRNOT00000011295
|
Gsg1
|
germ cell associated 1 |
| chr2_+_41442241 | 0.00 |
ENSRNOT00000067546
|
Pde4d
|
phosphodiesterase 4D |
| chr7_-_49741540 | 0.00 |
ENSRNOT00000006523
|
Myf6
|
myogenic factor 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Vsx1_Uncx_Prrx2_Shox2_Noto
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.6 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.3 | 2.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 0.7 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.2 | 1.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.4 | GO:0021918 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.1 | 0.6 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 0.3 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.1 | 1.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.2 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.1 | GO:0021550 | medulla oblongata development(GO:0021550) sensory system development(GO:0048880) |
| 0.0 | 0.2 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.3 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.4 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:1902276 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.7 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:1903015 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.0 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0060661 | mammary gland bud morphogenesis(GO:0060648) submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0021824 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) positive regulation of vascular wound healing(GO:0035470) glomerular endothelium development(GO:0072011) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.0 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:1901367 | response to L-cysteine(GO:1901367) |
| 0.0 | 0.1 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 0.2 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 0.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 2.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.3 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 6.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.0 | GO:0003922 | GMP synthase activity(GO:0003921) GMP synthase (glutamine-hydrolyzing) activity(GO:0003922) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 2.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |