Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
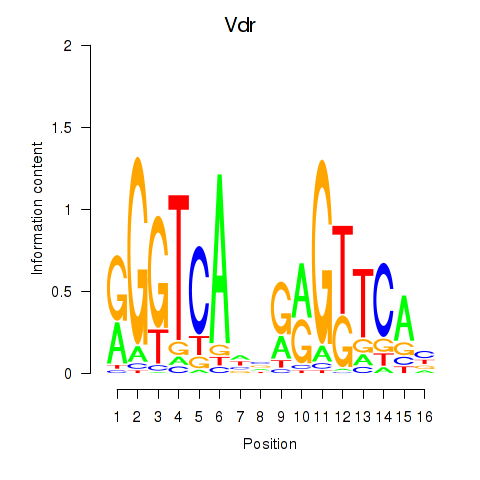
Results for Vdr
Z-value: 0.35
Transcription factors associated with Vdr
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Vdr
|
ENSRNOG00000054420 | vitamin D (1,25- dihydroxyvitamin D3) receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Vdr | rn6_v1_chr7_-_139394166_139394166 | 0.21 | 7.3e-01 | Click! |
Activity profile of Vdr motif
Sorted Z-values of Vdr motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_36239712 | 0.18 |
ENSRNOT00000071962
|
AABR07043711.2
|
|
| chr17_+_44794130 | 0.13 |
ENSRNOT00000077571
|
Hist1h2ac
|
histone cluster 1, H2ac |
| chr20_+_44680449 | 0.12 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr9_+_92435896 | 0.12 |
ENSRNOT00000022901
|
Fbxo36
|
F-box protein 36 |
| chr11_+_88122271 | 0.12 |
ENSRNOT00000002540
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr12_-_17186679 | 0.11 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr12_-_37047628 | 0.11 |
ENSRNOT00000001337
|
Rflna
|
refilin A |
| chr1_-_8751198 | 0.10 |
ENSRNOT00000030511
|
Adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr7_+_2458833 | 0.10 |
ENSRNOT00000085092
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr5_-_127983515 | 0.10 |
ENSRNOT00000043054
|
Fam159a
|
family with sequence similarity 159, member A |
| chr7_+_120153184 | 0.10 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chr10_+_99388130 | 0.10 |
ENSRNOT00000006238
|
Kcnj16
|
potassium voltage-gated channel subfamily J member 16 |
| chr1_+_99762253 | 0.09 |
ENSRNOT00000065876
|
Klk6
|
kallikrein related-peptidase 6 |
| chr16_+_61954590 | 0.09 |
ENSRNOT00000017883
|
Rbpms
|
RNA binding protein with multiple splicing |
| chr19_+_25946979 | 0.09 |
ENSRNOT00000004027
|
Gadd45gip1
|
GADD45G interacting protein 1 |
| chr9_+_100829552 | 0.09 |
ENSRNOT00000024695
|
Bok
|
BOK, BCL2 family apoptosis regulator |
| chr5_-_155252003 | 0.08 |
ENSRNOT00000017060
|
C1qb
|
complement C1q B chain |
| chr8_-_33661049 | 0.08 |
ENSRNOT00000068037
|
Fli1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_-_134638150 | 0.08 |
ENSRNOT00000013480
|
Tex38
|
testis expressed 38 |
| chr19_-_46167950 | 0.08 |
ENSRNOT00000071024
|
AABR07043892.1
|
|
| chr16_+_61954809 | 0.08 |
ENSRNOT00000068011
|
Rbpms
|
RNA binding protein with multiple splicing |
| chr7_+_143060597 | 0.08 |
ENSRNOT00000087481
|
Krt7
|
keratin 7 |
| chr5_+_135997052 | 0.08 |
ENSRNOT00000024921
|
Tctex1d4
|
Tctex1 domain containing 4 |
| chr12_+_23473270 | 0.08 |
ENSRNOT00000001935
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr1_+_198210525 | 0.08 |
ENSRNOT00000026755
|
Ypel3
|
yippee-like 3 |
| chr9_-_101107535 | 0.07 |
ENSRNOT00000080892
ENSRNOT00000071424 |
LOC100911516
|
coiled-coil-helix-coiled-coil-helix domain-containing protein 2, mitochondrial-like |
| chr7_+_2458264 | 0.07 |
ENSRNOT00000003550
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr5_+_152325871 | 0.07 |
ENSRNOT00000044711
|
Ftl1
|
ferritin light chain 1 |
| chr7_+_35125424 | 0.07 |
ENSRNOT00000085978
ENSRNOT00000010117 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr7_+_70821957 | 0.07 |
ENSRNOT00000045083
|
Ndufa4l2
|
NDUFA4, mitochondrial complex associated like 2 |
| chrX_-_15467875 | 0.07 |
ENSRNOT00000011207
|
Pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr16_+_21282467 | 0.07 |
ENSRNOT00000065345
|
Yjefn3
|
YjeF N-terminal domain containing 3 |
| chr4_-_38240848 | 0.07 |
ENSRNOT00000007567
|
Ndufa4
|
NADH:ubiquinone oxidoreductase subunit A4 |
| chr4_+_157726941 | 0.07 |
ENSRNOT00000025081
|
Vamp1
|
vesicle-associated membrane protein 1 |
| chr5_-_126782726 | 0.07 |
ENSRNOT00000048784
|
Tceanc2
|
transcription elongation factor A N-terminal and central domain containing 2 |
| chr20_+_4118461 | 0.07 |
ENSRNOT00000083085
|
RT1-Db2
|
RT1 class II, locus Db2 |
| chr2_-_257376756 | 0.07 |
ENSRNOT00000065811
|
Gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr14_+_46001849 | 0.06 |
ENSRNOT00000076611
|
Rell1
|
RELT-like 1 |
| chr9_+_100076958 | 0.06 |
ENSRNOT00000084297
|
Dusp28
|
dual specificity phosphatase 28 |
| chr7_+_2459141 | 0.06 |
ENSRNOT00000075681
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr19_-_10620671 | 0.06 |
ENSRNOT00000021842
|
Ccl17
|
C-C motif chemokine ligand 17 |
| chr3_+_162181974 | 0.06 |
ENSRNOT00000030203
|
Slc2a10
|
solute carrier family 2 member 10 |
| chr13_-_80738634 | 0.06 |
ENSRNOT00000081551
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr3_-_161294125 | 0.06 |
ENSRNOT00000021143
|
Spata25
|
spermatogenesis associated 25 |
| chr1_-_188190778 | 0.06 |
ENSRNOT00000092657
ENSRNOT00000022988 |
Coq7
|
coenzyme Q7, hydroxylase |
| chr16_+_9907598 | 0.06 |
ENSRNOT00000027352
|
Ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr1_-_53038229 | 0.06 |
ENSRNOT00000017282
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr1_-_220746224 | 0.06 |
ENSRNOT00000027734
|
Banf1
|
barrier to autointegration factor 1 |
| chr1_-_89559960 | 0.06 |
ENSRNOT00000092133
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chrX_+_105402867 | 0.06 |
ENSRNOT00000057222
|
Rpl36a
|
ribosomal protein L36a |
| chrX_+_112311251 | 0.06 |
ENSRNOT00000086698
|
AABR07040855.1
|
|
| chr1_-_224994653 | 0.06 |
ENSRNOT00000026313
|
Polr2g
|
RNA polymerase II subunit G |
| chr1_-_221507181 | 0.05 |
ENSRNOT00000093048
|
Arl2
|
ADP-ribosylation factor like GTPase 2 |
| chr15_+_31533675 | 0.05 |
ENSRNOT00000084141
|
AABR07017825.7
|
|
| chr1_+_197659187 | 0.05 |
ENSRNOT00000082228
|
LOC103690016
|
serine/threonine-protein kinase SBK1 |
| chr15_-_61564695 | 0.05 |
ENSRNOT00000068216
|
Rgcc
|
regulator of cell cycle |
| chr6_-_136145837 | 0.05 |
ENSRNOT00000015122
|
Ckb
|
creatine kinase B |
| chr5_+_129257429 | 0.05 |
ENSRNOT00000072396
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr5_-_157268903 | 0.05 |
ENSRNOT00000022716
|
Pla2g5
|
phospholipase A2, group V |
| chr12_+_22835019 | 0.05 |
ENSRNOT00000059530
|
Col26a1
|
collagen type XXVI alpha 1 chain |
| chrX_+_71272042 | 0.05 |
ENSRNOT00000076034
ENSRNOT00000076816 |
Gjb1
|
gap junction protein, beta 1 |
| chr8_+_62283336 | 0.05 |
ENSRNOT00000025410
|
Rpp25
|
ribonuclease P/MRP 25 subunit |
| chr8_+_22856539 | 0.05 |
ENSRNOT00000015381
|
Angptl8
|
angiopoietin-like 8 |
| chr1_-_80221710 | 0.05 |
ENSRNOT00000091687
|
Fosb
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr3_-_10153554 | 0.05 |
ENSRNOT00000093246
|
Exosc2
|
exosome component 2 |
| chr8_+_61901128 | 0.05 |
ENSRNOT00000033209
|
LOC691110
|
similar to taste receptor protein 1 |
| chr9_-_10054359 | 0.05 |
ENSRNOT00000072001
|
Clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr10_-_58924137 | 0.05 |
ENSRNOT00000066740
|
Tekt1
|
tektin 1 |
| chr3_-_71893600 | 0.04 |
ENSRNOT00000006787
ENSRNOT00000057685 |
Tfpi
|
tissue factor pathway inhibitor |
| chr16_+_20578131 | 0.04 |
ENSRNOT00000026713
|
Ssbp4
|
single stranded DNA binding protein 4 |
| chr1_-_103323476 | 0.04 |
ENSRNOT00000019051
|
Mrgprx3
|
MAS related GPR family member X3 |
| chr6_-_99274269 | 0.04 |
ENSRNOT00000058675
|
Tex21
|
testis expressed 21 |
| chr3_-_172566010 | 0.04 |
ENSRNOT00000071913
|
Atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr2_+_208577197 | 0.04 |
ENSRNOT00000076654
|
Wdr77
|
WD repeat domain 77 |
| chr10_-_87067456 | 0.04 |
ENSRNOT00000014163
|
Ccr7
|
C-C motif chemokine receptor 7 |
| chr6_+_99817431 | 0.04 |
ENSRNOT00000009621
|
Churc1
|
churchill domain containing 1 |
| chr7_-_130120579 | 0.04 |
ENSRNOT00000044376
|
Mapk12
|
mitogen-activated protein kinase 12 |
| chr18_+_31574821 | 0.04 |
ENSRNOT00000018499
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr18_+_24708115 | 0.04 |
ENSRNOT00000061054
|
Lims2
|
LIM zinc finger domain containing 2 |
| chr1_-_226732736 | 0.04 |
ENSRNOT00000072343
|
LOC108348129
|
pepsin F-like |
| chr11_-_60546997 | 0.04 |
ENSRNOT00000083124
ENSRNOT00000050092 |
Btla
|
B and T lymphocyte associated |
| chr17_+_20090246 | 0.04 |
ENSRNOT00000074600
|
Dtnbp1
|
dystrobrevin binding protein 1 |
| chr5_+_126783061 | 0.04 |
ENSRNOT00000013224
|
Tmem59
|
transmembrane protein 59 |
| chr5_+_144160108 | 0.04 |
ENSRNOT00000064972
|
Eva1b
|
eva-1 homolog B |
| chr12_+_41463922 | 0.04 |
ENSRNOT00000089378
|
Cfap73
|
cilia and flagella associated protein 73 |
| chr13_-_89518939 | 0.04 |
ENSRNOT00000004228
|
Sdhc
|
succinate dehydrogenase complex subunit C |
| chr13_+_77940454 | 0.04 |
ENSRNOT00000003460
|
Mrps14
|
mitochondrial ribosomal protein S14 |
| chr10_+_89166890 | 0.04 |
ENSRNOT00000088331
|
Ramp2
|
receptor activity modifying protein 2 |
| chr6_-_75551625 | 0.04 |
ENSRNOT00000005825
|
Sptssa
|
serine palmitoyltransferase, small subunit A |
| chr17_+_43661222 | 0.04 |
ENSRNOT00000022881
ENSRNOT00000022809 ENSRNOT00000022810 |
Hfe
|
hemochromatosis |
| chr10_+_13410899 | 0.04 |
ENSRNOT00000007392
ENSRNOT00000070981 |
Prss27
|
protease, serine 27 |
| chr10_-_14092289 | 0.04 |
ENSRNOT00000019624
|
Ndufb10
|
NADH:ubiquinone oxidoreductase subunit B10 |
| chr14_+_44524753 | 0.04 |
ENSRNOT00000076245
|
Rpl9
|
ribosomal protein L9 |
| chr12_+_41486076 | 0.04 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr12_+_12738812 | 0.03 |
ENSRNOT00000092233
ENSRNOT00000001379 |
Aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr19_-_11057254 | 0.03 |
ENSRNOT00000025559
|
Herpud1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr14_+_1463359 | 0.03 |
ENSRNOT00000070834
|
Csf2ra
|
colony stimulating factor 2 receptor alpha subunit |
| chr12_-_46920952 | 0.03 |
ENSRNOT00000001532
|
Msi1
|
musashi RNA-binding protein 1 |
| chr11_-_87240833 | 0.03 |
ENSRNOT00000052200
|
Tssk1b
|
testis-specific serine kinase 1B |
| chr3_+_175982777 | 0.03 |
ENSRNOT00000082073
|
Ntsr1
|
neurotensin receptor 1 |
| chr7_-_23843505 | 0.03 |
ENSRNOT00000006366
ENSRNOT00000077577 |
Fbxo7
|
F-box protein 7 |
| chr6_+_135866739 | 0.03 |
ENSRNOT00000013460
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr3_-_2680622 | 0.03 |
ENSRNOT00000020806
|
RGD1306215
|
similar to hypothetical protein MGC36831 |
| chrX_+_26294066 | 0.03 |
ENSRNOT00000037862
|
Hccs
|
holocytochrome c synthase |
| chr6_+_29977797 | 0.03 |
ENSRNOT00000071784
|
Fkbp1b
|
FK506 binding protein 1B |
| chr10_+_65767930 | 0.03 |
ENSRNOT00000039954
|
Vtn
|
vitronectin |
| chr3_-_172537877 | 0.03 |
ENSRNOT00000072069
|
Ctsz
|
cathepsin Z |
| chr13_+_80517536 | 0.03 |
ENSRNOT00000004386
|
Myoc
|
myocilin |
| chr1_-_189238776 | 0.03 |
ENSRNOT00000020817
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr20_-_12351813 | 0.03 |
ENSRNOT00000065236
|
Slc19a1
|
solute carrier family 19 member 1 |
| chr1_+_248723397 | 0.03 |
ENSRNOT00000072188
|
LOC100911854
|
mannose-binding protein C-like |
| chr18_-_15637715 | 0.03 |
ENSRNOT00000022270
|
Dsg2
|
desmoglein 2 |
| chr14_-_84662143 | 0.03 |
ENSRNOT00000057529
ENSRNOT00000080078 |
Hormad2
|
HORMA domain containing 2 |
| chr14_+_83724933 | 0.03 |
ENSRNOT00000029848
|
Pla2g3
|
phospholipase A2, group III |
| chr16_+_26859397 | 0.03 |
ENSRNOT00000044171
|
Msmo1
|
methylsterol monooxygenase 1 |
| chr19_+_58505811 | 0.03 |
ENSRNOT00000031717
|
Map10
|
microtubule-associated protein 10 |
| chr14_-_17225389 | 0.03 |
ENSRNOT00000052035
|
Art3
|
ADP-ribosyltransferase 3 |
| chr1_-_162385575 | 0.03 |
ENSRNOT00000016540
|
Thrsp
|
thyroid hormone responsive |
| chr16_-_83278119 | 0.03 |
ENSRNOT00000019454
|
Ing1
|
inhibitor of growth family, member 1 |
| chrX_+_110670119 | 0.03 |
ENSRNOT00000014811
ENSRNOT00000089579 |
LOC680663
|
hypothetical protein LOC680663 |
| chr19_-_26082719 | 0.03 |
ENSRNOT00000083159
|
Rnaseh2a
|
ribonuclease H2, subunit A |
| chr3_-_168111920 | 0.03 |
ENSRNOT00000046011
|
Cyp24a1
|
cytochrome P450, family 24, subfamily a, polypeptide 1 |
| chr5_-_157518511 | 0.03 |
ENSRNOT00000070865
ENSRNOT00000078028 |
Htr6
|
5-hydroxytryptamine receptor 6 |
| chr15_-_60512704 | 0.03 |
ENSRNOT00000049056
|
Tnfsf11
|
tumor necrosis factor superfamily member 11 |
| chr3_-_8751324 | 0.03 |
ENSRNOT00000030623
|
LOC499770
|
similar to LOC495800 protein |
| chr6_+_132551394 | 0.03 |
ENSRNOT00000019583
|
Evl
|
Enah/Vasp-like |
| chr4_+_152945760 | 0.03 |
ENSRNOT00000076032
|
Kdm5a
|
lysine demethylase 5A |
| chr5_+_159484370 | 0.03 |
ENSRNOT00000010593
|
Sdhb
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr4_-_98995590 | 0.03 |
ENSRNOT00000008698
|
Thnsl2
|
threonine synthase-like 2 |
| chr19_-_41541986 | 0.02 |
ENSRNOT00000059172
|
LOC100910419
|
transducin-like enhancer protein 2-like |
| chr12_-_22006533 | 0.02 |
ENSRNOT00000059593
|
LOC100362783
|
Uncharacterized protein C7orf61 homolog |
| chr1_+_102258124 | 0.02 |
ENSRNOT00000068434
|
Otog
|
otogelin |
| chr2_+_60180215 | 0.02 |
ENSRNOT00000084624
|
Prlr
|
prolactin receptor |
| chr5_+_137257637 | 0.02 |
ENSRNOT00000093001
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr1_-_165382216 | 0.02 |
ENSRNOT00000023648
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chr15_+_31462806 | 0.02 |
ENSRNOT00000072946
|
AABR07017825.3
|
|
| chr14_-_33150509 | 0.02 |
ENSRNOT00000002837
|
Rest
|
RE1-silencing transcription factor |
| chr1_+_140477868 | 0.02 |
ENSRNOT00000025008
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr1_-_141533908 | 0.02 |
ENSRNOT00000020117
|
Mesp1
|
mesoderm posterior bHLH transcription factor 1 |
| chr16_+_47368768 | 0.02 |
ENSRNOT00000017966
|
Wwc2
|
WW and C2 domain containing 2 |
| chrX_+_124894466 | 0.02 |
ENSRNOT00000080894
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr10_+_64952119 | 0.02 |
ENSRNOT00000012154
|
Pipox
|
pipecolic acid and sarcosine oxidase |
| chr10_-_63219309 | 0.02 |
ENSRNOT00000076357
|
Blmh
|
bleomycin hydrolase |
| chr4_+_149970237 | 0.02 |
ENSRNOT00000019529
|
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr3_-_11317049 | 0.02 |
ENSRNOT00000030558
|
Swi5
|
SWI5 homologous recombination repair protein |
| chr2_-_33920837 | 0.02 |
ENSRNOT00000075196
|
Erbin
|
erbb2 interacting protein |
| chr8_-_116422366 | 0.02 |
ENSRNOT00000023623
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr6_-_29999410 | 0.02 |
ENSRNOT00000075790
|
Sf3b6
|
splicing factor 3B, subunit 6 |
| chr5_-_138222534 | 0.02 |
ENSRNOT00000009691
|
Zfp691
|
zinc finger protein 691 |
| chr6_-_109103999 | 0.02 |
ENSRNOT00000009086
ENSRNOT00000080009 |
Acyp1
|
acylphosphatase 1 |
| chrX_+_28435507 | 0.02 |
ENSRNOT00000005615
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chrX_+_124894706 | 0.02 |
ENSRNOT00000066677
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr14_+_44524416 | 0.02 |
ENSRNOT00000038927
|
Rpl9
|
ribosomal protein L9 |
| chr4_+_149970567 | 0.02 |
ENSRNOT00000091765
|
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr1_+_61132818 | 0.02 |
ENSRNOT00000041434
|
Vom1r20
|
vomeronasal 1 receptor 20 |
| chr10_-_57822498 | 0.02 |
ENSRNOT00000037546
|
Nlrp1a
|
NLR family, pyrin domain containing 1A |
| chr1_+_57491643 | 0.02 |
ENSRNOT00000002038
|
Tbp
|
TATA box binding protein |
| chr16_+_9563218 | 0.02 |
ENSRNOT00000035915
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr4_+_133179309 | 0.02 |
ENSRNOT00000007419
|
Gxylt2
|
glucoside xylosyltransferase 2 |
| chr6_-_138093643 | 0.02 |
ENSRNOT00000045874
|
Igh-6
|
immunoglobulin heavy chain 6 |
| chr5_-_24631679 | 0.02 |
ENSRNOT00000010846
ENSRNOT00000067129 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr1_+_163398891 | 0.02 |
ENSRNOT00000020513
|
Gucy2e
|
guanylate cyclase 2E |
| chr1_-_71342633 | 0.02 |
ENSRNOT00000071400
|
Zscan5b
|
zinc finger and SCAN domain containing 5B |
| chr13_-_53086231 | 0.02 |
ENSRNOT00000036094
|
LOC498236
|
LRRGT00186 |
| chr3_-_114235933 | 0.02 |
ENSRNOT00000023939
|
Duox2
|
dual oxidase 2 |
| chr1_+_193335645 | 0.02 |
ENSRNOT00000019640
|
Lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr3_+_151335292 | 0.02 |
ENSRNOT00000073642
|
Mmp24
|
matrix metallopeptidase 24 |
| chr9_+_77320726 | 0.02 |
ENSRNOT00000068450
|
Spag16
|
sperm associated antigen 16 |
| chr13_+_99335020 | 0.02 |
ENSRNOT00000029787
|
AABR07021930.1
|
|
| chr12_+_25561777 | 0.02 |
ENSRNOT00000002032
|
Rcc1l
|
RCC1 like |
| chr10_-_48044344 | 0.02 |
ENSRNOT00000003697
|
Prpsap2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chrX_+_123751089 | 0.02 |
ENSRNOT00000092384
|
Nkap
|
NFKB activating protein |
| chr16_+_50111306 | 0.02 |
ENSRNOT00000019302
|
Cyp4v3
|
cytochrome P450, family 4, subfamily v, polypeptide 3 |
| chr16_-_18645941 | 0.02 |
ENSRNOT00000079241
|
Dydc2
|
DPY30 domain containing 2 |
| chr20_+_40769586 | 0.02 |
ENSRNOT00000001079
|
Fabp7
|
fatty acid binding protein 7 |
| chr4_+_148589144 | 0.02 |
ENSRNOT00000044870
|
Olr827
|
olfactory receptor 827 |
| chr1_-_98493978 | 0.02 |
ENSRNOT00000023942
|
Nkg7
|
natural killer cell granule protein 7 |
| chr17_+_24015982 | 0.02 |
ENSRNOT00000024122
|
Nol7
|
nucleolar protein 7 |
| chr14_-_44524252 | 0.02 |
ENSRNOT00000003779
|
Lias
|
lipoic acid synthetase |
| chrM_+_7006 | 0.02 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr8_-_116092763 | 0.02 |
ENSRNOT00000020781
|
Hemk1
|
HemK methyltransferase family member 1 |
| chr19_-_53625665 | 0.02 |
ENSRNOT00000079651
ENSRNOT00000068764 |
Fbxo31
|
F-box protein 31 |
| chr18_-_58423196 | 0.02 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr7_+_36826360 | 0.02 |
ENSRNOT00000029783
|
Ndufa13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| chr5_-_158313426 | 0.02 |
ENSRNOT00000025488
|
Pax7
|
paired box 7 |
| chr3_+_111013856 | 0.02 |
ENSRNOT00000016854
|
Zfyve19
|
zinc finger FYVE-type containing 19 |
| chr4_-_183697531 | 0.02 |
ENSRNOT00000055441
|
Amn1
|
antagonist of mitotic exit network 1 homolog |
| chr16_+_6609668 | 0.02 |
ENSRNOT00000021862
|
Tkt
|
transketolase |
| chr2_+_200076054 | 0.02 |
ENSRNOT00000025327
|
Sec22b
|
SEC22 homolog B, vesicle trafficking protein |
| chr9_-_69960086 | 0.02 |
ENSRNOT00000072737
|
Gpr1
|
G protein-coupled receptor 1 |
| chr19_-_26084679 | 0.02 |
ENSRNOT00000046181
|
Rnaseh2a
|
ribonuclease H2, subunit A |
| chr15_-_34566719 | 0.02 |
ENSRNOT00000064643
|
Sdr39u1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr12_+_48101673 | 0.02 |
ENSRNOT00000065060
|
Foxn4
|
forkhead box N4 |
| chr16_-_81693000 | 0.02 |
ENSRNOT00000092353
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr5_+_126670825 | 0.02 |
ENSRNOT00000012201
|
Cyb5rl
|
cytochrome b5 reductase-like |
| chr1_+_83744238 | 0.02 |
ENSRNOT00000028249
|
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr13_+_91768256 | 0.02 |
ENSRNOT00000071206
|
LOC681470
|
similar to olfactory receptor 1403 |
| chr16_-_74662194 | 0.02 |
ENSRNOT00000034670
|
Tpte2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr9_-_98551410 | 0.01 |
ENSRNOT00000066346
ENSRNOT00000086678 |
Hes6
|
hes family bHLH transcription factor 6 |
| chr15_-_27997810 | 0.01 |
ENSRNOT00000034647
|
Olr1637
|
olfactory receptor 1637 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Vdr
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.1 | GO:1904708 | granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.0 | GO:0071049 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.0 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.0 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.0 | 0.0 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.0 | 0.0 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.0 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.0 | 0.0 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070821 | azurophil granule membrane(GO:0035577) tertiary granule membrane(GO:0070821) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0045281 | respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |