Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
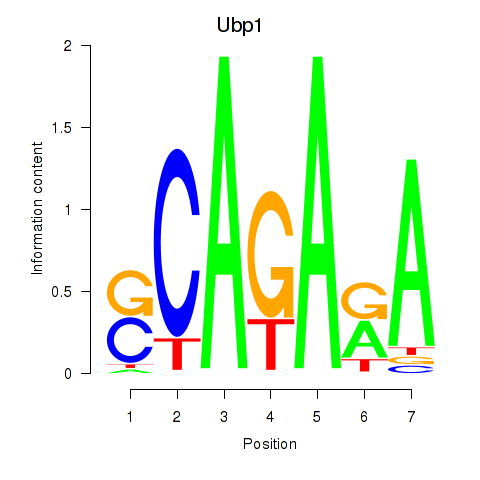
Results for Ubp1
Z-value: 0.36
Transcription factors associated with Ubp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ubp1
|
ENSRNOG00000009729 | upstream binding protein 1 (LBP-1a) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ubp1 | rn6_v1_chr8_+_122197027_122197027 | -0.89 | 4.1e-02 | Click! |
Activity profile of Ubp1 motif
Sorted Z-values of Ubp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_53678777 | 0.21 |
ENSRNOT00000045944
|
Drd2
|
dopamine receptor D2 |
| chr2_-_113616766 | 0.19 |
ENSRNOT00000016858
ENSRNOT00000074723 |
Tmem212
|
transmembrane protein 212 |
| chr10_-_70871066 | 0.16 |
ENSRNOT00000015139
|
Ccl3
|
C-C motif chemokine ligand 3 |
| chr14_+_43694183 | 0.15 |
ENSRNOT00000046342
|
RGD1563570
|
similar to ribosomal protein S23 |
| chrX_-_32153794 | 0.15 |
ENSRNOT00000005348
|
Tmem27
|
transmembrane protein 27 |
| chr1_+_168957460 | 0.12 |
ENSRNOT00000090745
|
LOC103694857
|
hemoglobin subunit beta-2 |
| chr2_+_189993262 | 0.12 |
ENSRNOT00000016034
|
S100a3
|
S100 calcium binding protein A3 |
| chr9_-_94250809 | 0.11 |
ENSRNOT00000026387
|
Ecel1
|
endothelin converting enzyme-like 1 |
| chr12_+_19714324 | 0.11 |
ENSRNOT00000072303
|
RGD1559588
|
similar to cell surface receptor FDFACT |
| chr15_+_17834635 | 0.11 |
ENSRNOT00000085530
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr4_+_70828894 | 0.11 |
ENSRNOT00000064892
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr2_+_127538659 | 0.11 |
ENSRNOT00000093483
ENSRNOT00000058476 |
Slc25a31
|
solute carrier family 25 member 31 |
| chr12_-_2007516 | 0.10 |
ENSRNOT00000037564
|
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr1_-_213650247 | 0.09 |
ENSRNOT00000019679
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb |
| chr18_+_63130542 | 0.09 |
ENSRNOT00000024947
|
Tubb6
|
tubulin, beta 6 class V |
| chr10_+_14828597 | 0.09 |
ENSRNOT00000025434
|
Tekt4
|
tektin 4 |
| chr7_+_120153184 | 0.09 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chr8_+_67753279 | 0.09 |
ENSRNOT00000009716
|
Calml4
|
calmodulin-like 4 |
| chr2_+_197655786 | 0.08 |
ENSRNOT00000028732
|
Ctss
|
cathepsin S |
| chr5_+_2043583 | 0.08 |
ENSRNOT00000082813
|
Eloc
|
elongin C |
| chr6_+_49825469 | 0.08 |
ENSRNOT00000006921
|
Fam150b
|
family with sequence similarity 150, member B |
| chr10_-_13168217 | 0.08 |
ENSRNOT00000087768
|
Elob
|
elongin B |
| chr6_+_99817431 | 0.08 |
ENSRNOT00000009621
|
Churc1
|
churchill domain containing 1 |
| chr3_-_60813869 | 0.08 |
ENSRNOT00000058234
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr19_-_26082719 | 0.07 |
ENSRNOT00000083159
|
Rnaseh2a
|
ribonuclease H2, subunit A |
| chr3_+_4282236 | 0.07 |
ENSRNOT00000042201
|
LOC100364457
|
ribosomal protein L9-like |
| chr8_+_58431407 | 0.07 |
ENSRNOT00000011974
|
Sln
|
sarcolipin |
| chr1_+_83003841 | 0.07 |
ENSRNOT00000057384
|
Ceacam4
|
carcinoembryonic antigen-related cell adhesion molecule 4 |
| chr17_+_87020089 | 0.07 |
ENSRNOT00000043523
|
Rpl37-ps1
|
ribosomal protein L37, pseudogene 1 |
| chr18_-_29562153 | 0.07 |
ENSRNOT00000023977
|
Cd14
|
CD14 molecule |
| chr5_-_61410812 | 0.07 |
ENSRNOT00000015270
|
Igfbpl1
|
insulin-like growth factor binding protein-like 1 |
| chr9_+_10535340 | 0.07 |
ENSRNOT00000075408
|
Znrf4
|
zinc and ring finger 4 |
| chr7_-_123586919 | 0.07 |
ENSRNOT00000011484
|
Ndufa6
|
NADH:ubiquinone oxidoreductase subunit A6 |
| chr11_+_70967439 | 0.07 |
ENSRNOT00000032518
|
Iqcg
|
IQ motif containing G |
| chr20_-_32353677 | 0.07 |
ENSRNOT00000035355
|
Stox1
|
storkhead box 1 |
| chr19_-_39267928 | 0.07 |
ENSRNOT00000027686
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr11_-_88177448 | 0.07 |
ENSRNOT00000085475
|
Ypel1
|
yippee-like 1 |
| chr3_+_138174054 | 0.07 |
ENSRNOT00000007946
|
Banf2
|
barrier to autointegration factor 2 |
| chr7_-_115979298 | 0.07 |
ENSRNOT00000075883
|
RGD1308195
|
similar to secreted Ly6/uPAR related protein 2 |
| chr16_+_83161880 | 0.07 |
ENSRNOT00000080793
|
1700016D06Rik
|
RIKEN cDNA 1700016D06 gene |
| chr3_+_91870208 | 0.07 |
ENSRNOT00000040249
|
Rpl21
|
ribosomal protein L21 |
| chr18_+_63394900 | 0.07 |
ENSRNOT00000024126
|
Psmg2
|
proteasome assembly chaperone 2 |
| chr6_+_26602144 | 0.06 |
ENSRNOT00000008037
|
Ucn
|
urocortin |
| chr9_+_15582564 | 0.06 |
ENSRNOT00000020640
|
LOC100912849
|
uncharacterized LOC100912849 |
| chr2_+_225645568 | 0.06 |
ENSRNOT00000017878
|
Abca4
|
ATP binding cassette subfamily A member 4 |
| chr12_-_20486276 | 0.06 |
ENSRNOT00000074057
|
LOC680910
|
similar to paired immunoglobin-like type 2 receptor beta |
| chr4_+_181243338 | 0.06 |
ENSRNOT00000034051
|
Smco2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr1_-_77535681 | 0.06 |
ENSRNOT00000018559
|
Selenow
|
selenoprotein W |
| chr11_-_83483513 | 0.06 |
ENSRNOT00000084380
|
AABR07034673.1
|
|
| chr11_+_31002451 | 0.06 |
ENSRNOT00000002838
|
Eva1c
|
eva-1 homolog C |
| chr9_+_66335492 | 0.06 |
ENSRNOT00000037555
|
RGD1562029
|
similar to KIAA2012 protein |
| chr1_-_80783898 | 0.06 |
ENSRNOT00000045306
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr15_+_61879184 | 0.06 |
ENSRNOT00000042606
|
Sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr4_-_50312608 | 0.06 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr4_-_89695928 | 0.06 |
ENSRNOT00000039316
|
Gprin3
|
GPRIN family member 3 |
| chr7_-_18577325 | 0.06 |
ENSRNOT00000084308
ENSRNOT00000090849 |
March2
|
membrane associated ring-CH-type finger 2 |
| chr10_-_77512032 | 0.06 |
ENSRNOT00000003295
|
Pctp
|
phosphatidylcholine transfer protein |
| chr8_+_99568958 | 0.06 |
ENSRNOT00000073400
|
Plscr5
|
phospholipid scramblase family, member 5 |
| chr6_+_69619295 | 0.06 |
ENSRNOT00000089761
|
AABR07064218.1
|
|
| chr2_+_208749996 | 0.06 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr11_+_77815181 | 0.06 |
ENSRNOT00000002640
|
Cldn1
|
claudin 1 |
| chr8_+_70994563 | 0.05 |
ENSRNOT00000051504
ENSRNOT00000077163 |
Spg21
|
spastic paraplegia 21 homolog (human) |
| chr1_-_89084859 | 0.05 |
ENSRNOT00000032026
|
Cox6b1
|
cytochrome c oxidase subunit 6B1 |
| chr8_-_19917396 | 0.05 |
ENSRNOT00000032963
|
Olr1158
|
olfactory receptor 1158 |
| chr3_+_93351619 | 0.05 |
ENSRNOT00000011035
|
Elf5
|
E74-like factor 5 |
| chr5_-_118541928 | 0.05 |
ENSRNOT00000012947
|
Itgb3bp
|
integrin subunit beta 3 binding protein |
| chr1_+_282063929 | 0.05 |
ENSRNOT00000041410
|
AABR07007121.1
|
|
| chr7_-_80796670 | 0.05 |
ENSRNOT00000010539
|
Abra
|
actin-binding Rho activating protein |
| chrX_-_156440461 | 0.05 |
ENSRNOT00000083951
|
Rpl10
|
ribosomal protein L10 |
| chr3_+_112242270 | 0.05 |
ENSRNOT00000080533
ENSRNOT00000082876 |
Capn3
|
calpain 3 |
| chr18_-_55859333 | 0.05 |
ENSRNOT00000025989
|
Myoz3
|
myozenin 3 |
| chr20_+_33945829 | 0.05 |
ENSRNOT00000064063
|
LOC103689994
|
radial spoke head protein 4 homolog A |
| chr14_-_85484275 | 0.05 |
ENSRNOT00000083770
|
Kremen1
|
kringle containing transmembrane protein 1 |
| chr9_+_67774150 | 0.05 |
ENSRNOT00000091060
|
Icos
|
inducible T-cell co-stimulator |
| chr1_-_222468896 | 0.05 |
ENSRNOT00000028754
|
Cox8a
|
cytochrome c oxidase subunit 8A |
| chr5_-_58019836 | 0.05 |
ENSRNOT00000066977
|
Enho
|
energy homeostasis associated |
| chr19_+_37282018 | 0.05 |
ENSRNOT00000021723
|
Tmem208
|
transmembrane protein 208 |
| chr20_+_32450733 | 0.05 |
ENSRNOT00000036449
|
Rsph4a
|
radial spoke head 4 homolog A |
| chr11_+_34557679 | 0.05 |
ENSRNOT00000002289
|
Ripply3
|
ripply transcriptional repressor 3 |
| chr1_-_212568224 | 0.05 |
ENSRNOT00000024976
|
LOC100911225
|
fucose mutarotase-like |
| chr5_-_139933764 | 0.05 |
ENSRNOT00000015278
|
LOC680700
|
similar to ribosomal protein L10a |
| chr7_+_114997103 | 0.05 |
ENSRNOT00000010224
|
Ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr2_+_140708397 | 0.05 |
ENSRNOT00000088846
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr4_-_101393329 | 0.05 |
ENSRNOT00000007636
|
RGD1562515
|
similar to RIKEN cDNA 4931417E11 |
| chr19_+_39357791 | 0.05 |
ENSRNOT00000086435
ENSRNOT00000015006 |
Cyb5b
|
cytochrome b5 type B |
| chr1_+_99616447 | 0.05 |
ENSRNOT00000029197
|
Klk14
|
kallikrein related-peptidase 14 |
| chr8_-_128521109 | 0.05 |
ENSRNOT00000034025
|
Scn11a
|
sodium voltage-gated channel alpha subunit 11 |
| chr12_-_18540166 | 0.05 |
ENSRNOT00000001792
|
Il3ra
|
interleukin 3 receptor subunit alpha |
| chr4_-_89151184 | 0.05 |
ENSRNOT00000010263
|
Nap1l5
|
nucleosome assembly protein 1-like 5 |
| chr1_-_266086299 | 0.05 |
ENSRNOT00000026609
|
Cuedc2
|
CUE domain containing 2 |
| chr5_-_167999853 | 0.05 |
ENSRNOT00000087402
|
Park7
|
Parkinsonism associated deglycase |
| chr1_+_72580424 | 0.05 |
ENSRNOT00000022977
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chrX_-_72078551 | 0.05 |
ENSRNOT00000076978
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr3_-_2853272 | 0.05 |
ENSRNOT00000023022
|
Fcna
|
ficolin A |
| chr2_-_197981665 | 0.05 |
ENSRNOT00000083086
|
Mrps21
|
mitochondrial ribosomal protein S21 |
| chr9_-_43127887 | 0.05 |
ENSRNOT00000021685
|
Ankrd39
|
ankyrin repeat domain 39 |
| chr10_-_56412544 | 0.05 |
ENSRNOT00000020578
|
Tmem102
|
transmembrane protein 102 |
| chr3_-_52998650 | 0.05 |
ENSRNOT00000009988
|
RGD1560831
|
similar to 40S ribosomal protein S3 |
| chr1_-_89179825 | 0.05 |
ENSRNOT00000028514
|
Tmem147
|
transmembrane protein 147 |
| chr5_+_152466331 | 0.05 |
ENSRNOT00000082841
|
LOC108351043
|
uncharacterized LOC108351043 |
| chr4_-_152835182 | 0.05 |
ENSRNOT00000036721
|
B4galnt3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr17_+_5311274 | 0.04 |
ENSRNOT00000067020
|
LOC102547665
|
spermatogenesis-associated protein 31D1-like |
| chr13_-_32427177 | 0.04 |
ENSRNOT00000044628
|
Cdh19
|
cadherin 19 |
| chr1_+_224933529 | 0.04 |
ENSRNOT00000088282
|
Wdr74
|
WD repeat domain 74 |
| chr17_+_69960160 | 0.04 |
ENSRNOT00000023887
|
Ucn3
|
urocortin 3 |
| chr9_-_4168221 | 0.04 |
ENSRNOT00000061861
|
Sult1c2a
|
sulfotransferase family, cytosolic, 1C, member 2a |
| chr2_-_211017778 | 0.04 |
ENSRNOT00000026883
|
Sypl2
|
synaptophysin-like 2 |
| chr3_-_122006803 | 0.04 |
ENSRNOT00000089660
|
RGD1566226
|
similar to hypothetical protein F830045P16 |
| chr1_+_141832774 | 0.04 |
ENSRNOT00000073302
|
Fuom
|
fucose mutarotase |
| chr1_+_224933920 | 0.04 |
ENSRNOT00000066823
|
Wdr74
|
WD repeat domain 74 |
| chr16_-_21017163 | 0.04 |
ENSRNOT00000027661
|
Mef2b
|
myocyte enhancer factor 2B |
| chrX_+_28539158 | 0.04 |
ENSRNOT00000073535
|
Tlr8
|
toll-like receptor 8 |
| chr7_+_27248115 | 0.04 |
ENSRNOT00000031946
ENSRNOT00000059545 |
LOC362863
|
first gene upstream of Nt5dc3 |
| chr9_+_88918433 | 0.04 |
ENSRNOT00000021730
|
Ccl20
|
C-C motif chemokine ligand 20 |
| chr1_+_89408935 | 0.04 |
ENSRNOT00000075068
|
LOC100909893
|
protein FAM187B-like |
| chr17_+_41798783 | 0.04 |
ENSRNOT00000023519
|
Nrsn1
|
neurensin 1 |
| chr1_+_220869805 | 0.04 |
ENSRNOT00000015962
|
Cfl1
|
cofilin 1 |
| chrX_+_31984612 | 0.04 |
ENSRNOT00000005181
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr1_-_201110928 | 0.04 |
ENSRNOT00000027721
|
Nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr6_-_26820959 | 0.04 |
ENSRNOT00000043572
|
Khk
|
ketohexokinase |
| chr2_+_187415262 | 0.04 |
ENSRNOT00000056923
|
Gpatch4
|
G patch domain containing 4 |
| chr20_-_5227620 | 0.04 |
ENSRNOT00000086240
|
RT1-DMb
|
RT1 class II, locus DMb |
| chr2_+_202200797 | 0.04 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr4_-_70996395 | 0.04 |
ENSRNOT00000021485
|
Kel
|
Kell blood group, metallo-endopeptidase |
| chr20_+_1809078 | 0.04 |
ENSRNOT00000084853
|
Olr1739
|
olfactory receptor 1739 |
| chr10_+_68588789 | 0.04 |
ENSRNOT00000049614
|
AABR07030122.1
|
|
| chr6_+_136150785 | 0.04 |
ENSRNOT00000078211
ENSRNOT00000015308 |
LOC103692719
|
tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A |
| chr2_+_238529074 | 0.04 |
ENSRNOT00000016195
|
Ppa2
|
pyrophosphatase (inorganic) 2 |
| chr6_+_41917132 | 0.04 |
ENSRNOT00000071387
|
Ntsr2
|
neurotensin receptor 2 |
| chr7_-_98804499 | 0.04 |
ENSRNOT00000070909
|
Tatdn1
|
TatD DNase domain containing 1 |
| chr10_-_63219309 | 0.04 |
ENSRNOT00000076357
|
Blmh
|
bleomycin hydrolase |
| chr16_-_71809925 | 0.04 |
ENSRNOT00000022425
|
Tm2d2
|
TM2 domain containing 2 |
| chr4_+_78735279 | 0.04 |
ENSRNOT00000011970
|
Malsu1
|
mitochondrial assembly of ribosomal large subunit 1 |
| chr7_+_77966722 | 0.04 |
ENSRNOT00000006142
|
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr12_+_3969493 | 0.04 |
ENSRNOT00000042692
|
Vom2r59
|
vomeronasal 2 receptor, 59 |
| chr10_-_104163634 | 0.04 |
ENSRNOT00000005198
|
Mif4gd
|
MIF4G domain containing |
| chr11_-_31805728 | 0.04 |
ENSRNOT00000032162
|
Gart
|
phosphoribosylglycinamide formyltransferase |
| chr20_+_17750744 | 0.04 |
ENSRNOT00000000745
|
RGD1559903
|
similar to RIKEN cDNA 1700049L16 |
| chr7_+_140054592 | 0.04 |
ENSRNOT00000014337
|
Olr1108
|
olfactory receptor 1108 |
| chr13_-_98023829 | 0.04 |
ENSRNOT00000075426
|
Kif28p
|
kinesin family member 28, pseudogene |
| chr7_-_124982566 | 0.04 |
ENSRNOT00000075099
|
Sult4a1
|
sulfotransferase family 4A, member 1 |
| chr19_-_10212050 | 0.04 |
ENSRNOT00000082822
|
Tepp
|
testis, prostate and placenta expressed |
| chr5_-_152247332 | 0.04 |
ENSRNOT00000077165
|
Lin28a
|
lin-28 homolog A |
| chr1_-_101236065 | 0.04 |
ENSRNOT00000066834
|
Cd37
|
CD37 molecule |
| chr4_+_61850348 | 0.04 |
ENSRNOT00000013423
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr13_+_42220251 | 0.04 |
ENSRNOT00000078185
|
AABR07020815.1
|
|
| chr2_+_154556029 | 0.04 |
ENSRNOT00000084238
|
Gmps
|
guanine monophosphate synthase |
| chr4_+_14151343 | 0.04 |
ENSRNOT00000061687
ENSRNOT00000076573 ENSRNOT00000077219 ENSRNOT00000008319 |
Cd36
|
CD36 molecule |
| chr7_+_143060597 | 0.04 |
ENSRNOT00000087481
|
Krt7
|
keratin 7 |
| chr9_+_27343853 | 0.04 |
ENSRNOT00000072794
|
Tmem14a
|
transmembrane protein 14A |
| chr2_-_210726179 | 0.04 |
ENSRNOT00000025689
|
Gstm7
|
glutathione S-transferase, mu 7 |
| chrX_+_77885586 | 0.04 |
ENSRNOT00000034329
|
RGD1560821
|
similar to small nuclear ribonucleoparticle-associated protein |
| chr1_+_64162672 | 0.04 |
ENSRNOT00000089713
|
Tfpt
|
TCF3 (E2A) fusion partner |
| chr10_-_57436368 | 0.04 |
ENSRNOT00000056608
|
Scimp
|
SLP adaptor and CSK interacting membrane protein |
| chr3_+_113415774 | 0.04 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr14_-_91996774 | 0.04 |
ENSRNOT00000005851
|
Ddc
|
dopa decarboxylase |
| chr14_-_18849258 | 0.04 |
ENSRNOT00000033406
|
Pf4
|
platelet factor 4 |
| chr15_+_4850122 | 0.04 |
ENSRNOT00000071133
|
Gng2
|
G protein subunit gamma 2 |
| chr4_+_144382945 | 0.04 |
ENSRNOT00000007601
|
Cav3
|
caveolin 3 |
| chr10_+_89635675 | 0.04 |
ENSRNOT00000028179
|
RGD1561590
|
similar to SAP18 |
| chr19_+_52374298 | 0.04 |
ENSRNOT00000092131
|
Atp2c2
|
ATPase secretory pathway Ca2+ transporting 2 |
| chr12_+_12649861 | 0.04 |
ENSRNOT00000092402
|
Ccz1b
|
CCZ1 homolog B, vacuolar protein trafficking and biogenesis associated |
| chr13_+_89919667 | 0.04 |
ENSRNOT00000006196
|
Itln1
|
intelectin 1 |
| chr8_-_133128290 | 0.04 |
ENSRNOT00000008783
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1 |
| chr1_+_94718402 | 0.04 |
ENSRNOT00000046035
|
Uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr4_-_82702429 | 0.04 |
ENSRNOT00000011069
|
Hibadh
|
3-hydroxyisobutyrate dehydrogenase |
| chr18_+_81821127 | 0.04 |
ENSRNOT00000058199
|
Fbxo15
|
F-box protein 15 |
| chr10_-_63491279 | 0.04 |
ENSRNOT00000082301
|
Tusc5
|
tumor suppressor candidate 5 |
| chr20_-_11556312 | 0.04 |
ENSRNOT00000091856
|
RGD1561557
|
similar to chromosome 21 open reading frame 29 |
| chr2_-_23289266 | 0.04 |
ENSRNOT00000061708
|
Bhmt2
|
betaine-homocysteine S-methyltransferase 2 |
| chr1_-_53014466 | 0.04 |
ENSRNOT00000049831
|
Sft2d1
|
SFT2 domain containing 1 |
| chr1_-_141655417 | 0.03 |
ENSRNOT00000068084
|
Ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr2_-_252451999 | 0.03 |
ENSRNOT00000021861
|
Dnase2b
|
deoxyribonuclease 2 beta |
| chr15_-_28044210 | 0.03 |
ENSRNOT00000033809
|
Rnase1l1
|
ribonuclease, RNase A family, 1-like 1 (pancreatic) |
| chr10_+_35680658 | 0.03 |
ENSRNOT00000004111
|
Mrnip
|
MRN complex interacting protein |
| chr5_+_133865331 | 0.03 |
ENSRNOT00000035409
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr1_-_8751198 | 0.03 |
ENSRNOT00000030511
|
Adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr2_+_193627243 | 0.03 |
ENSRNOT00000082934
|
AABR07012331.1
|
|
| chrX_-_105355716 | 0.03 |
ENSRNOT00000015043
|
Timm8a1
|
translocase of inner mitochondrial membrane 8 homolog A1 (yeast) |
| chr7_-_107203897 | 0.03 |
ENSRNOT00000086263
|
Lrrc6
|
leucine rich repeat containing 6 |
| chrX_-_915953 | 0.03 |
ENSRNOT00000075264
|
Spaca5
|
sperm acrosome associated 5 |
| chr7_+_141249044 | 0.03 |
ENSRNOT00000084911
|
Aqp5
|
aquaporin 5 |
| chr16_+_55834458 | 0.03 |
ENSRNOT00000086141
|
AABR07025928.1
|
|
| chr17_-_11037738 | 0.03 |
ENSRNOT00000043796
|
AABR07027015.1
|
|
| chr16_-_47207487 | 0.03 |
ENSRNOT00000059459
|
Dctd
|
dCMP deaminase |
| chr12_-_6879154 | 0.03 |
ENSRNOT00000001207
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr11_+_70967223 | 0.03 |
ENSRNOT00000087633
|
Iqcg
|
IQ motif containing G |
| chr4_-_23135354 | 0.03 |
ENSRNOT00000011432
|
Steap4
|
STEAP4 metalloreductase |
| chr12_+_37829579 | 0.03 |
ENSRNOT00000084587
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr8_+_56451240 | 0.03 |
ENSRNOT00000085278
|
LOC100359687
|
mitochondrial ribosomal protein L1-like |
| chr9_+_95274707 | 0.03 |
ENSRNOT00000045163
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr1_+_216293087 | 0.03 |
ENSRNOT00000027875
ENSRNOT00000087153 |
Kcnq1
|
potassium voltage-gated channel subfamily Q member 1 |
| chr15_-_34612432 | 0.03 |
ENSRNOT00000090206
|
Mcpt1l1
|
mast cell protease 1-like 1 |
| chr7_+_35125424 | 0.03 |
ENSRNOT00000085978
ENSRNOT00000010117 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr11_-_31892531 | 0.03 |
ENSRNOT00000033745
|
Cryzl1
|
crystallin zeta like 1 |
| chr4_+_32495321 | 0.03 |
ENSRNOT00000015286
|
Sdhaf3
|
succinate dehydrogenase complex assembly factor 3 |
| chr2_+_128675814 | 0.03 |
ENSRNOT00000058366
|
RGD1359508
|
similar to protein C33A12.3 |
| chr20_+_5040337 | 0.03 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chr12_+_19680712 | 0.03 |
ENSRNOT00000081310
|
AABR07035561.2
|
|
| chr8_+_28045093 | 0.03 |
ENSRNOT00000073373
|
Thyn1
|
thymocyte nuclear protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ubp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1900166 | glial cell-derived neurotrophic factor secretion(GO:0044467) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) negative regulation of dopamine receptor signaling pathway(GO:0060160) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.0 | 0.1 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.0 | GO:0043132 | NAD transport(GO:0043132) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) response to butyrate(GO:1903544) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.1 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.1 | GO:0035482 | gastric motility(GO:0035482) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.0 | GO:1903200 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) TRAIL receptor biosynthetic process(GO:0045557) regulation of TRAIL receptor biosynthetic process(GO:0045560) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.0 | 0.2 | GO:0071223 | response to lipoteichoic acid(GO:0070391) cellular response to lipoteichoic acid(GO:0071223) |
| 0.0 | 0.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.0 | GO:2000769 | regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.0 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.0 | GO:1901367 | response to L-cysteine(GO:1901367) |
| 0.0 | 0.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.0 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.0 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.0 | GO:0044299 | C-fiber(GO:0044299) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.0 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.0 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.0 | GO:0003922 | GMP synthase activity(GO:0003921) GMP synthase (glutamine-hydrolyzing) activity(GO:0003922) |
| 0.0 | 0.0 | GO:0004058 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.0 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |