Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
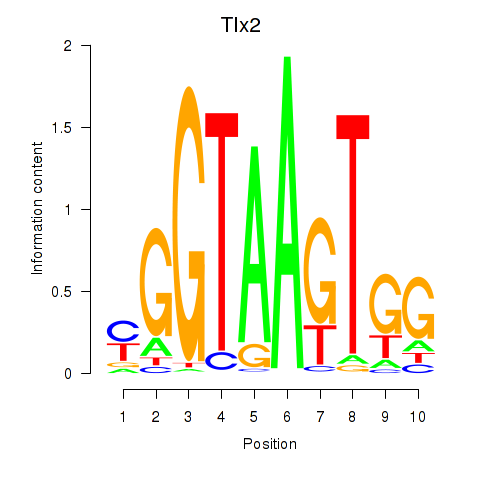
Results for Tlx2
Z-value: 0.28
Transcription factors associated with Tlx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tlx2
|
ENSRNOG00000055188 | T-cell leukemia homeobox 2 |
|
Tlx2
|
ENSRNOG00000061571 | T-cell leukemia homeobox 2 |
Activity profile of Tlx2 motif
Sorted Z-values of Tlx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_19714324 | 0.17 |
ENSRNOT00000072303
|
RGD1559588
|
similar to cell surface receptor FDFACT |
| chr17_-_90315492 | 0.14 |
ENSRNOT00000070807
|
Gng4
|
G protein subunit gamma 4 |
| chr1_+_276659542 | 0.13 |
ENSRNOT00000019681
|
Tcf7l2
|
transcription factor 7 like 2 |
| chr11_+_31389514 | 0.11 |
ENSRNOT00000000325
|
Olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr5_-_17061361 | 0.10 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr2_-_190100276 | 0.10 |
ENSRNOT00000015351
|
S100a9
|
S100 calcium binding protein A9 |
| chr3_+_147585947 | 0.09 |
ENSRNOT00000006833
|
Scrt2
|
scratch family transcriptional repressor 2 |
| chr19_-_43911057 | 0.09 |
ENSRNOT00000026017
|
Ctrb1
|
chymotrypsinogen B1 |
| chr15_+_41069507 | 0.08 |
ENSRNOT00000018533
|
C1qtnf9
|
C1q and tumor necrosis factor related protein 9 |
| chr5_-_34813116 | 0.08 |
ENSRNOT00000017479
|
Nkain3
|
Sodium/potassium transporting ATPase interacting 3 |
| chr7_-_73216251 | 0.08 |
ENSRNOT00000057587
|
LOC100362027
|
ribosomal protein L30-like |
| chr14_+_5928737 | 0.08 |
ENSRNOT00000071877
ENSRNOT00000040985 ENSRNOT00000074889 |
Mpa2l
|
macrophage activation 2 like |
| chr4_+_165732643 | 0.08 |
ENSRNOT00000034403
|
LOC690326
|
hypothetical protein LOC690326 |
| chr7_-_12646960 | 0.08 |
ENSRNOT00000014687
|
Prtn3
|
proteinase 3 |
| chr8_-_66863476 | 0.08 |
ENSRNOT00000018820
|
Rplp1
|
ribosomal protein lateral stalk subunit P1 |
| chr2_+_199199845 | 0.08 |
ENSRNOT00000020932
|
LOC100909441
|
tubulin alpha-1C chain-like |
| chr13_-_80738634 | 0.07 |
ENSRNOT00000081551
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr5_-_136762986 | 0.07 |
ENSRNOT00000026852
|
Artn
|
artemin |
| chr13_-_90814119 | 0.07 |
ENSRNOT00000011208
|
Tagln2
|
transgelin 2 |
| chr9_+_94928489 | 0.07 |
ENSRNOT00000087366
ENSRNOT00000085770 ENSRNOT00000024733 |
Sag
|
S-antigen visual arrestin |
| chr1_+_81643816 | 0.07 |
ENSRNOT00000027214
|
LOC103689942
|
carcinoembryonic antigen-related cell adhesion molecule 1-like |
| chr5_+_145257714 | 0.07 |
ENSRNOT00000019214
|
Dlgap3
|
DLG associated protein 3 |
| chr8_-_63092009 | 0.07 |
ENSRNOT00000034300
|
Loxl1
|
lysyl oxidase-like 1 |
| chr10_+_70655818 | 0.06 |
ENSRNOT00000074839
|
LOC689039
|
hypothetical protein LOC689039 |
| chr2_-_260596777 | 0.06 |
ENSRNOT00000009370
|
Lhx8
|
LIM homeobox 8 |
| chr10_-_40805941 | 0.06 |
ENSRNOT00000017588
|
Atox1
|
antioxidant 1 copper chaperone |
| chr7_+_123168811 | 0.06 |
ENSRNOT00000007091
|
Csdc2
|
cold shock domain containing C2 |
| chr8_+_71914867 | 0.06 |
ENSRNOT00000023372
|
Dapk2
|
death-associated protein kinase 2 |
| chr15_-_41691518 | 0.06 |
ENSRNOT00000020036
|
Ebpl
|
emopamil binding protein-like |
| chr9_+_84689150 | 0.06 |
ENSRNOT00000072242
|
Kcne4
|
potassium voltage-gated channel subfamily E regulatory subunit 4 |
| chr20_-_4921348 | 0.06 |
ENSRNOT00000082497
ENSRNOT00000041151 |
RT1-CE4
|
RT1 class I, locus CE4 |
| chr7_-_121232741 | 0.06 |
ENSRNOT00000023196
|
Pdgfb
|
platelet derived growth factor subunit B |
| chr12_+_19890749 | 0.06 |
ENSRNOT00000074970
|
RGD1559588
|
similar to cell surface receptor FDFACT |
| chr4_+_98371184 | 0.06 |
ENSRNOT00000086911
|
AABR07060872.1
|
|
| chr9_-_101388833 | 0.06 |
ENSRNOT00000071817
|
Chchd2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr4_+_168832910 | 0.06 |
ENSRNOT00000011134
|
Gprc5a
|
G protein-coupled receptor, class C, group 5, member A |
| chr20_+_5351605 | 0.05 |
ENSRNOT00000089306
ENSRNOT00000041590 ENSRNOT00000081240 ENSRNOT00000082538 |
RT1-A1
|
RT1 class Ia, locus A1 |
| chr8_+_44847157 | 0.05 |
ENSRNOT00000080288
|
Clmp
|
CXADR-like membrane protein |
| chr10_-_103590607 | 0.05 |
ENSRNOT00000034741
|
Cd300le
|
Cd300 molecule-like family member E |
| chr19_+_561727 | 0.05 |
ENSRNOT00000016259
ENSRNOT00000081547 |
Rrad
|
RRAD, Ras related glycolysis inhibitor and calcium channel regulator |
| chr3_+_13838304 | 0.05 |
ENSRNOT00000025067
|
Hspa5
|
heat shock protein family A member 5 |
| chr7_-_2712723 | 0.05 |
ENSRNOT00000004363
|
Il23a
|
interleukin 23 subunit alpha |
| chr10_-_66020682 | 0.05 |
ENSRNOT00000011019
|
Fam58b
|
family with sequence similarity 58, member B |
| chr3_+_147042944 | 0.05 |
ENSRNOT00000012608
|
Fkbp1a
|
FK506 binding protein 1a |
| chr2_+_187347602 | 0.05 |
ENSRNOT00000025384
|
Nes
|
nestin |
| chr11_+_67082193 | 0.05 |
ENSRNOT00000003129
|
Cd86
|
CD86 molecule |
| chr1_+_88113445 | 0.05 |
ENSRNOT00000056955
|
Ggn
|
gametogenetin |
| chr5_-_62025152 | 0.05 |
ENSRNOT00000042649
|
Hemgn
|
hemogen |
| chr8_-_83280888 | 0.05 |
ENSRNOT00000052341
|
Gfral
|
GDNF family receptor alpha like |
| chr4_+_83391283 | 0.05 |
ENSRNOT00000031365
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr5_-_128009735 | 0.05 |
ENSRNOT00000013913
|
Gpx7
|
glutathione peroxidase 7 |
| chr8_+_62925357 | 0.05 |
ENSRNOT00000011074
ENSRNOT00000090588 |
Stra6
|
stimulated by retinoic acid 6 |
| chr4_+_86275717 | 0.05 |
ENSRNOT00000016414
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr1_-_224986291 | 0.05 |
ENSRNOT00000026284
|
Taf6l
|
TATA-box binding protein associated factor 6 like |
| chr2_+_190073815 | 0.05 |
ENSRNOT00000015473
|
S100a8
|
S100 calcium binding protein A8 |
| chr2_-_183128932 | 0.05 |
ENSRNOT00000031179
|
Mnd1
|
meiotic nuclear divisions 1 |
| chr1_-_190596431 | 0.05 |
ENSRNOT00000079335
|
Pdzd9
|
PDZ domain containing 9 |
| chr18_-_55771730 | 0.05 |
ENSRNOT00000026426
|
Smim3
|
small integral membrane protein 3 |
| chr4_+_62780596 | 0.05 |
ENSRNOT00000073846
|
LOC689574
|
hypothetical protein LOC689574 |
| chr10_-_37099004 | 0.04 |
ENSRNOT00000075465
|
N4bp3
|
Nedd4 binding protein 3 |
| chr2_+_203768729 | 0.04 |
ENSRNOT00000018895
|
Igsf3
|
immunoglobulin superfamily, member 3 |
| chr12_+_11191812 | 0.04 |
ENSRNOT00000033537
|
Atp5j2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2 |
| chr7_-_118933812 | 0.04 |
ENSRNOT00000031951
|
Apol9a
|
apolipoprotein L 9a |
| chr5_-_148577292 | 0.04 |
ENSRNOT00000017156
|
Zcchc17
|
zinc finger CCHC-type containing 17 |
| chr2_+_189955836 | 0.04 |
ENSRNOT00000078351
|
S100a3
|
S100 calcium binding protein A3 |
| chr1_-_80271001 | 0.04 |
ENSRNOT00000034266
|
Cd3eap
|
CD3e molecule associated protein |
| chr20_-_5754773 | 0.04 |
ENSRNOT00000035977
|
Ip6k3
|
inositol hexakisphosphate kinase 3 |
| chr2_+_200397967 | 0.04 |
ENSRNOT00000025821
|
Reg4
|
regenerating family member 4 |
| chr1_-_153740905 | 0.04 |
ENSRNOT00000023239
|
Prss23
|
protease, serine, 23 |
| chr13_+_49003803 | 0.04 |
ENSRNOT00000080556
|
Lemd1
|
LEM domain containing 1 |
| chr15_+_23553128 | 0.04 |
ENSRNOT00000012985
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr10_-_35005287 | 0.04 |
ENSRNOT00000032386
|
LOC103689949
|
nedd4 binding protein 3 |
| chr18_-_27520295 | 0.04 |
ENSRNOT00000027544
|
Gfra3
|
GDNF family receptor alpha 3 |
| chr15_-_6587367 | 0.04 |
ENSRNOT00000038449
|
Zfp385d
|
zinc finger protein 385D |
| chr19_-_55967836 | 0.04 |
ENSRNOT00000051452
|
Sult5a1
|
sulfotransferase family 5A, member 1 |
| chr2_+_233602732 | 0.04 |
ENSRNOT00000044232
|
Pitx2
|
paired-like homeodomain 2 |
| chr1_-_221087449 | 0.04 |
ENSRNOT00000016987
|
Fam89b
|
family with sequence similarity 89, member B |
| chr10_+_47490153 | 0.04 |
ENSRNOT00000003182
|
Aldh3a1
|
aldehyde dehydrogenase 3 family, member A1 |
| chrX_-_139916883 | 0.04 |
ENSRNOT00000090442
|
Gpc3
|
glypican 3 |
| chr16_+_6035926 | 0.04 |
ENSRNOT00000020284
|
Selenok
|
selenoprotein K |
| chr15_+_31948035 | 0.04 |
ENSRNOT00000071627
|
AABR07017868.1
|
|
| chr15_+_33069382 | 0.04 |
ENSRNOT00000060193
|
Mrpl52
|
mitochondrial ribosomal protein L52 |
| chr7_+_122160171 | 0.04 |
ENSRNOT00000074499
|
LOC100362980
|
CG3918-like |
| chr19_-_914880 | 0.04 |
ENSRNOT00000017127
|
Cklf
|
chemokine-like factor |
| chr7_-_29233392 | 0.04 |
ENSRNOT00000064241
|
Spic
|
Spi-C transcription factor |
| chr4_+_66670618 | 0.04 |
ENSRNOT00000010796
|
Tbxas1
|
thromboxane A synthase 1 |
| chr4_+_7257752 | 0.03 |
ENSRNOT00000088357
|
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr11_-_81660395 | 0.03 |
ENSRNOT00000048739
|
Fetub
|
fetuin B |
| chr4_-_158704170 | 0.03 |
ENSRNOT00000082009
|
Ntf3
|
neurotrophin 3 |
| chr4_+_130332076 | 0.03 |
ENSRNOT00000083127
|
Mitf
|
melanogenesis associated transcription factor |
| chr19_-_25288335 | 0.03 |
ENSRNOT00000050214
|
Nanos3
|
nanos C2HC-type zinc finger 3 |
| chr10_+_3227160 | 0.03 |
ENSRNOT00000088075
|
Ntan1
|
N-terminal asparagine amidase |
| chr12_+_21678580 | 0.03 |
ENSRNOT00000039797
|
LOC100910669
|
paired immunoglobulin-like type 2 receptor alpha-like |
| chr3_-_170955399 | 0.03 |
ENSRNOT00000084990
|
Bmp7
|
bone morphogenetic protein 7 |
| chr2_+_115678344 | 0.03 |
ENSRNOT00000036717
|
Slc7a14
|
solute carrier family 7, member 14 |
| chr5_-_130085838 | 0.03 |
ENSRNOT00000035252
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr14_-_18733391 | 0.03 |
ENSRNOT00000003745
|
Cxcl2
|
C-X-C motif chemokine ligand 2 |
| chr3_-_161812243 | 0.03 |
ENSRNOT00000025238
ENSRNOT00000087646 |
Slc35c2
|
solute carrier family 35 member C2 |
| chr7_+_140716113 | 0.03 |
ENSRNOT00000033450
|
Tuba1c
|
tubulin, alpha 1C |
| chr16_-_20592449 | 0.03 |
ENSRNOT00000026821
|
Isyna1
|
inositol-3-phosphate synthase 1 |
| chr4_+_140703619 | 0.03 |
ENSRNOT00000009563
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr5_+_79179417 | 0.03 |
ENSRNOT00000010454
|
Orm1
|
orosomucoid 1 |
| chr4_+_25435873 | 0.03 |
ENSRNOT00000000018
|
Steap1
|
STEAP family member 1 |
| chr11_-_32508420 | 0.03 |
ENSRNOT00000002717
|
Kcne1
|
potassium voltage-gated channel subfamily E regulatory subunit 1 |
| chr6_+_126170720 | 0.03 |
ENSRNOT00000065246
|
Rin3
|
Ras and Rab interactor 3 |
| chr10_+_14598014 | 0.03 |
ENSRNOT00000068336
|
Tsr3
|
TSR3, 20S rRNA accumulation |
| chr10_-_56850085 | 0.03 |
ENSRNOT00000025767
|
Rnasek
|
ribonuclease K |
| chr3_-_111941841 | 0.03 |
ENSRNOT00000010701
|
Ehd4
|
EH-domain containing 4 |
| chr5_+_159606647 | 0.03 |
ENSRNOT00000051693
|
Rps21-ps1
|
ribosomal protein S21, pseudogene 1 |
| chr9_+_67763897 | 0.03 |
ENSRNOT00000071226
|
Icos
|
inducible T-cell co-stimulator |
| chr15_+_31533675 | 0.03 |
ENSRNOT00000084141
|
AABR07017825.7
|
|
| chr8_-_40137390 | 0.03 |
ENSRNOT00000042717
|
Panx3
|
pannexin 3 |
| chr5_-_57896475 | 0.03 |
ENSRNOT00000017903
|
RGD1561916
|
similar to testes development-related NYD-SP22 isoform 1 |
| chr9_+_43331155 | 0.03 |
ENSRNOT00000023036
|
Zap70
|
zeta chain of T cell receptor associated protein kinase 70 |
| chr2_+_239415046 | 0.03 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr9_+_24066303 | 0.03 |
ENSRNOT00000018163
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr6_+_126170911 | 0.03 |
ENSRNOT00000077477
|
Rin3
|
Ras and Rab interactor 3 |
| chr3_-_9738752 | 0.03 |
ENSRNOT00000045993
|
Ptges
|
prostaglandin E synthase |
| chr10_+_106800142 | 0.03 |
ENSRNOT00000074899
|
LOC688282
|
hypothetical protein LOC688282 |
| chr10_-_103972668 | 0.03 |
ENSRNOT00000004836
|
Atp5h
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit d |
| chr12_-_9988618 | 0.03 |
ENSRNOT00000087449
|
Rasl11a
|
RAS-like family 11 member A |
| chr10_-_64202380 | 0.03 |
ENSRNOT00000008982
|
Rflnb
|
refilin B |
| chr10_-_71383378 | 0.03 |
ENSRNOT00000077025
|
Dusp14
|
dual specificity phosphatase 14 |
| chr8_+_116857684 | 0.03 |
ENSRNOT00000026711
|
Mst1
|
macrophage stimulating 1 |
| chr1_-_88112683 | 0.03 |
ENSRNOT00000090615
|
Spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr10_+_37215937 | 0.03 |
ENSRNOT00000006567
|
Sar1b
|
secretion associated, Ras related GTPase 1B |
| chrX_+_122808605 | 0.03 |
ENSRNOT00000017567
|
Zcchc12
|
zinc finger CCHC-type containing 12 |
| chr17_+_76079720 | 0.03 |
ENSRNOT00000073933
|
Proser2
|
proline and serine rich 2 |
| chr2_+_119139717 | 0.03 |
ENSRNOT00000016051
|
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr15_-_28517335 | 0.03 |
ENSRNOT00000060514
ENSRNOT00000090378 |
LOC100911576
|
heterogeneous nuclear ribonucleoproteins C1/C2-like |
| chr2_-_88113029 | 0.03 |
ENSRNOT00000013354
|
Car2
|
carbonic anhydrase 2 |
| chr9_-_82699551 | 0.03 |
ENSRNOT00000020673
|
Obsl1
|
obscurin-like 1 |
| chr1_-_87155118 | 0.03 |
ENSRNOT00000072441
|
AABR07002854.1
|
|
| chr1_-_90208036 | 0.03 |
ENSRNOT00000076255
ENSRNOT00000076967 |
Lsm14a
|
LSM14A mRNA processing body assembly factor |
| chr11_+_46009322 | 0.03 |
ENSRNOT00000002233
|
Tmem45a
|
transmembrane protein 45A |
| chrX_+_113948654 | 0.03 |
ENSRNOT00000068431
|
Tmem164
|
transmembrane protein 164 |
| chr4_-_97784842 | 0.03 |
ENSRNOT00000007698
|
Gadd45a
|
growth arrest and DNA-damage-inducible, alpha |
| chr1_-_52544450 | 0.03 |
ENSRNOT00000043474
|
Pde10a
|
phosphodiesterase 10A |
| chr5_+_139038134 | 0.03 |
ENSRNOT00000078155
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr9_-_94250809 | 0.03 |
ENSRNOT00000026387
|
Ecel1
|
endothelin converting enzyme-like 1 |
| chr11_+_67451549 | 0.03 |
ENSRNOT00000042777
|
Stfa2l2
|
stefin A2-like 2 |
| chr13_-_112099336 | 0.03 |
ENSRNOT00000009158
ENSRNOT00000044161 |
Camk1g
|
calcium/calmodulin-dependent protein kinase IG |
| chr11_+_46714355 | 0.03 |
ENSRNOT00000073404
|
Tmem45al
|
transmembrane protein 45A-like |
| chr12_-_9998779 | 0.03 |
ENSRNOT00000001265
|
Rpl21
|
ribosomal protein L21 |
| chr1_-_101054851 | 0.03 |
ENSRNOT00000027849
|
Prrg2
|
proline rich and Gla domain 2 |
| chr4_-_147742114 | 0.03 |
ENSRNOT00000041464
|
Efcab12
|
EF-hand calcium binding domain 12 |
| chr6_-_108488330 | 0.03 |
ENSRNOT00000016076
|
Npc2
|
NPC intracellular cholesterol transporter 2 |
| chr6_+_108488678 | 0.03 |
ENSRNOT00000016089
|
Isca2
|
iron-sulfur cluster assembly 2 |
| chr5_-_154394328 | 0.03 |
ENSRNOT00000030043
|
Rpl11
|
ribosomal protein L11 |
| chr2_-_123193130 | 0.03 |
ENSRNOT00000019554
|
Anxa5
|
annexin A5 |
| chr19_-_15570611 | 0.02 |
ENSRNOT00000022679
|
Mmp2
|
matrix metallopeptidase 2 |
| chr3_-_176465162 | 0.02 |
ENSRNOT00000048807
|
Nkain4
|
Sodium/potassium transporting ATPase interacting 4 |
| chr15_+_33596301 | 0.02 |
ENSRNOT00000022563
|
Il25
|
interleukin 25 |
| chr2_+_187282000 | 0.02 |
ENSRNOT00000065211
|
Hdgf
|
hepatoma-derived growth factor |
| chr14_-_2032593 | 0.02 |
ENSRNOT00000000037
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr19_-_10653800 | 0.02 |
ENSRNOT00000022128
|
Cx3cl1
|
C-X3-C motif chemokine ligand 1 |
| chr6_+_22302069 | 0.02 |
ENSRNOT00000039132
|
Dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr1_-_64021321 | 0.02 |
ENSRNOT00000090819
|
Rps9
|
ribosomal protein S9 |
| chr7_+_14589520 | 0.02 |
ENSRNOT00000060210
|
Cyp4f37
|
cytochrome P450, family 4, subfamily f, polypeptide 37 |
| chr4_+_8166082 | 0.02 |
ENSRNOT00000076429
|
Srpk2
|
SRSF protein kinase 2 |
| chr20_+_12917111 | 0.02 |
ENSRNOT00000038953
|
Ybey
|
ybeY metallopeptidase |
| chr3_-_114307250 | 0.02 |
ENSRNOT00000064592
|
Shf
|
Src homology 2 domain containing F |
| chr1_+_264827739 | 0.02 |
ENSRNOT00000021553
|
Kazald1
|
Kazal-type serine peptidase inhibitor domain 1 |
| chr20_-_5110191 | 0.02 |
ENSRNOT00000076597
|
Csnk2b
|
casein kinase 2 beta |
| chr1_-_89017290 | 0.02 |
ENSRNOT00000028438
|
Psenen
|
presenilin enhancer gamma secretase subunit |
| chr1_+_198120099 | 0.02 |
ENSRNOT00000073652
|
Bola2
|
bolA family member 2 |
| chr8_+_94686938 | 0.02 |
ENSRNOT00000013285
|
Ripply2
|
ripply transcriptional repressor 2 |
| chr20_+_44521279 | 0.02 |
ENSRNOT00000085987
|
Fyn
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr13_-_53870428 | 0.02 |
ENSRNOT00000000812
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr3_+_170252901 | 0.02 |
ENSRNOT00000005871
|
Mc3r
|
melanocortin 3 receptor |
| chr11_+_45751812 | 0.02 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr10_+_89352835 | 0.02 |
ENSRNOT00000028060
|
Rpl27
|
ribosomal protein L27 |
| chr4_+_61850348 | 0.02 |
ENSRNOT00000013423
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr8_+_48443767 | 0.02 |
ENSRNOT00000010127
|
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr1_+_198655742 | 0.02 |
ENSRNOT00000023944
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr2_+_248398917 | 0.02 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr7_+_74047814 | 0.02 |
ENSRNOT00000014879
|
Osr2
|
odd-skipped related transciption factor 2 |
| chr3_+_148150698 | 0.02 |
ENSRNOT00000087075
ENSRNOT00000010251 |
Hm13
|
histocompatibility minor 13 |
| chr7_+_70349863 | 0.02 |
ENSRNOT00000031796
|
Cdk4
|
cyclin-dependent kinase 4 |
| chr7_-_107963073 | 0.02 |
ENSRNOT00000093353
ENSRNOT00000010980 |
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr7_-_18180705 | 0.02 |
ENSRNOT00000065954
|
Zfp494
|
zinc finger protein 494 |
| chr4_+_149908375 | 0.02 |
ENSRNOT00000019504
|
LOC100909657
|
uncharacterized LOC100909657 |
| chr2_-_149347796 | 0.02 |
ENSRNOT00000091020
|
P2ry14
|
purinergic receptor P2Y14 |
| chr1_-_187766670 | 0.02 |
ENSRNOT00000092513
ENSRNOT00000092282 |
Rps15a
|
ribosomal protein S15a |
| chr5_-_154394023 | 0.02 |
ENSRNOT00000079823
|
Rpl11
|
ribosomal protein L11 |
| chr6_+_34030620 | 0.02 |
ENSRNOT00000091651
|
Laptm4a
|
lysosomal protein transmembrane 4 alpha |
| chr1_-_86923575 | 0.02 |
ENSRNOT00000027029
|
Mrps12
|
mitochondrial ribosomal protein S12 |
| chr8_-_39266959 | 0.02 |
ENSRNOT00000046590
|
Ei24
|
EI24, autophagy associated transmembrane protein |
| chr19_-_41029206 | 0.02 |
ENSRNOT00000023383
|
Vac14
|
Vac14, PIKFYVE complex component |
| chr15_+_42522108 | 0.02 |
ENSRNOT00000021621
|
Ccdc25
|
coiled-coil domain containing 25 |
| chrX_-_112328642 | 0.02 |
ENSRNOT00000083150
|
Psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr17_-_18828929 | 0.02 |
ENSRNOT00000051732
|
LOC102554838
|
stathmin domain-containing protein 1-like |
| chr10_+_43871613 | 0.02 |
ENSRNOT00000080540
ENSRNOT00000003797 |
LOC24906
|
RoBo-1 |
| chr7_+_116632506 | 0.02 |
ENSRNOT00000009811
|
Gpihbp1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr10_+_62303662 | 0.02 |
ENSRNOT00000056355
|
Tlcd2
|
TLC domain containing 2 |
| chr8_-_14156656 | 0.02 |
ENSRNOT00000089364
|
Deup1
|
deuterosome assembly protein 1 |
| chr4_+_129574264 | 0.02 |
ENSRNOT00000010185
|
Arl6ip5
|
ADP-ribosylation factor like GTPase 6 interacting protein 5 |
| chr10_+_86860685 | 0.02 |
ENSRNOT00000084644
|
Rara
|
retinoic acid receptor, alpha |
| chr11_+_58624198 | 0.02 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chr1_+_89017479 | 0.02 |
ENSRNOT00000038154
|
U2af1l4
|
U2 small nuclear RNA auxiliary factor 1-like 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tlx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.1 | GO:0072254 | metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) |
| 0.0 | 0.1 | GO:1904401 | cellular response to Thyroid stimulating hormone(GO:1904401) |
| 0.0 | 0.1 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 0.0 | 0.1 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.1 | GO:0051140 | positive regulation of memory T cell differentiation(GO:0043382) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.0 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.0 | 0.0 | GO:0046865 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0045105 | intermediate filament polymerization or depolymerization(GO:0045105) |
| 0.0 | 0.0 | GO:0021539 | pulmonary myocardium development(GO:0003350) subthalamus development(GO:0021539) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.0 | GO:0072011 | glomerular endothelium development(GO:0072011) |
| 0.0 | 0.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.0 | GO:0072134 | nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 0.0 | 0.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.0 | GO:0036023 | embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.0 | GO:0043366 | beta selection(GO:0043366) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |