Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
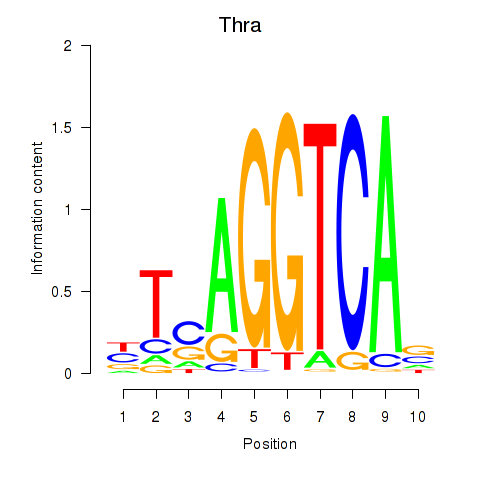
Results for Thra
Z-value: 0.22
Transcription factors associated with Thra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Thra
|
ENSRNOG00000009066 | thyroid hormone receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Thra | rn6_v1_chr10_+_86657285_86657285 | 0.65 | 2.3e-01 | Click! |
Activity profile of Thra motif
Sorted Z-values of Thra motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_115616766 | 0.10 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chr3_+_95715193 | 0.08 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr20_+_3827367 | 0.08 |
ENSRNOT00000079967
|
Rxrb
|
retinoid X receptor beta |
| chr1_+_82174451 | 0.07 |
ENSRNOT00000027783
|
Tmem145
|
transmembrane protein 145 |
| chr10_+_14547172 | 0.06 |
ENSRNOT00000092043
|
Unkl
|
unkempt family like zinc finger |
| chr5_+_147069616 | 0.05 |
ENSRNOT00000072908
|
Trim62
|
tripartite motif-containing 62 |
| chr8_-_61919874 | 0.05 |
ENSRNOT00000064830
|
RGD1305464
|
similar to human chromosome 15 open reading frame 39 |
| chr6_+_110624856 | 0.05 |
ENSRNOT00000014017
|
Vash1
|
vasohibin 1 |
| chr14_+_104250617 | 0.05 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chrX_-_159841072 | 0.05 |
ENSRNOT00000001158
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr3_+_80676820 | 0.05 |
ENSRNOT00000084809
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr4_+_153385205 | 0.04 |
ENSRNOT00000016561
|
Bcl2l13
|
BCL2 like 13 |
| chr7_-_12424367 | 0.04 |
ENSRNOT00000060698
|
Midn
|
midnolin |
| chr20_+_5008508 | 0.04 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr6_+_112203679 | 0.03 |
ENSRNOT00000031205
|
Nrxn3
|
neurexin 3 |
| chr11_+_64601029 | 0.03 |
ENSRNOT00000004138
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr8_-_47339343 | 0.03 |
ENSRNOT00000081007
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr1_-_214414763 | 0.03 |
ENSRNOT00000025240
ENSRNOT00000077776 |
LOC100911440
|
mitochondrial glutamate carrier 1-like |
| chr6_+_146784915 | 0.03 |
ENSRNOT00000008362
|
Sp8
|
Sp8 transcription factor |
| chr8_+_116332796 | 0.03 |
ENSRNOT00000021408
|
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr1_+_1784078 | 0.03 |
ENSRNOT00000020098
|
Lats1
|
large tumor suppressor kinase 1 |
| chr13_-_35668968 | 0.03 |
ENSRNOT00000042862
ENSRNOT00000003428 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr5_+_116421894 | 0.03 |
ENSRNOT00000080577
ENSRNOT00000086628 ENSRNOT00000004017 |
Nfia
|
nuclear factor I/A |
| chr6_-_135829953 | 0.03 |
ENSRNOT00000080623
ENSRNOT00000039059 |
Cdc42bpb
|
CDC42 binding protein kinase beta |
| chr10_+_72486152 | 0.03 |
ENSRNOT00000004193
|
RGD1310166
|
similar to Chromodomain-helicase-DNA-binding protein 1 (CHD-1) |
| chr10_-_59762644 | 0.03 |
ENSRNOT00000083390
|
Ctns
|
cystinosin, lysosomal cystine transporter |
| chr5_-_57632177 | 0.02 |
ENSRNOT00000080787
ENSRNOT00000092581 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr15_-_55209342 | 0.02 |
ENSRNOT00000021752
|
Rb1
|
RB transcriptional corepressor 1 |
| chr7_-_33916338 | 0.02 |
ENSRNOT00000005492
|
Cfap54
|
cilia and flagella associated protein 54 |
| chr16_+_19044607 | 0.02 |
ENSRNOT00000085462
|
Med26
|
mediator complex subunit 26 |
| chr12_+_24761210 | 0.02 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr5_+_113725717 | 0.02 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chrX_-_157095274 | 0.02 |
ENSRNOT00000084088
|
Abcd1
|
ATP binding cassette subfamily D member 1 |
| chr10_+_59000397 | 0.02 |
ENSRNOT00000021134
|
Mybbp1a
|
MYB binding protein 1a |
| chr3_-_2727616 | 0.02 |
ENSRNOT00000061904
|
C8g
|
complement C8 gamma chain |
| chr1_+_88922433 | 0.02 |
ENSRNOT00000045212
|
Nphs1
|
NPHS1 nephrin |
| chr5_-_160219812 | 0.02 |
ENSRNOT00000016333
|
Plekhm2
|
pleckstrin homology and RUN domain containing M2 |
| chrX_+_68752597 | 0.02 |
ENSRNOT00000077039
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr9_+_111597037 | 0.02 |
ENSRNOT00000021758
|
Fer
|
FER tyrosine kinase |
| chr11_+_84745904 | 0.02 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr10_-_18443934 | 0.02 |
ENSRNOT00000059895
ENSRNOT00000080021 |
Ranbp17
|
RAN binding protein 17 |
| chr14_+_12218553 | 0.02 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr1_-_66212418 | 0.02 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr2_+_198312428 | 0.02 |
ENSRNOT00000028758
|
Sf3b4
|
splicing factor 3b, subunit 4 |
| chr5_+_14415841 | 0.02 |
ENSRNOT00000010682
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr1_+_224824799 | 0.02 |
ENSRNOT00000024757
|
Slc22a6
|
solute carrier family 22 member 6 |
| chr17_+_36334147 | 0.02 |
ENSRNOT00000050261
|
E2f3
|
E2F transcription factor 3 |
| chr4_-_145948996 | 0.02 |
ENSRNOT00000043476
|
Atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr1_-_64446818 | 0.02 |
ENSRNOT00000081980
|
Myadm
|
myeloid-associated differentiation marker |
| chr3_+_148510779 | 0.01 |
ENSRNOT00000012156
|
Xkr7
|
XK related 7 |
| chr7_-_122926336 | 0.01 |
ENSRNOT00000000205
|
Chadl
|
chondroadherin-like |
| chr17_+_77167740 | 0.01 |
ENSRNOT00000042881
|
Optn
|
optineurin |
| chr7_-_142180997 | 0.01 |
ENSRNOT00000087632
|
Tfcp2
|
transcription factor CP2 |
| chr6_+_29106141 | 0.01 |
ENSRNOT00000084560
|
Atad2b
|
ATPase family, AAA domain containing 2B |
| chr6_+_2216623 | 0.01 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr10_-_85435016 | 0.01 |
ENSRNOT00000079921
|
4933428G20Rik
|
RIKEN cDNA 4933428G20 gene |
| chr7_-_140437467 | 0.01 |
ENSRNOT00000087181
|
Fkbp11
|
FK506 binding protein 11 |
| chr1_-_134867001 | 0.01 |
ENSRNOT00000090149
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr3_+_150055749 | 0.01 |
ENSRNOT00000055335
|
Actl10
|
actin-like 10 |
| chr1_+_177048655 | 0.01 |
ENSRNOT00000081595
|
Mical2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr10_+_85608544 | 0.01 |
ENSRNOT00000092122
|
Mllt6
|
MLLT6, PHD finger domain containing |
| chr8_+_50559126 | 0.01 |
ENSRNOT00000024918
|
Apoa5
|
apolipoprotein A5 |
| chr5_-_137238354 | 0.01 |
ENSRNOT00000039235
|
Szt2
|
seizure threshold 2 homolog (mouse) |
| chr14_-_83219071 | 0.01 |
ENSRNOT00000085788
|
Depdc5
|
DEP domain containing 5 |
| chr5_-_152473868 | 0.01 |
ENSRNOT00000022130
|
Fam110d
|
family with sequence similarity 110, member D |
| chr5_-_147691997 | 0.01 |
ENSRNOT00000011887
|
Tssk3
|
testis-specific serine kinase 3 |
| chr11_+_73936750 | 0.01 |
ENSRNOT00000002350
|
Atp13a3
|
ATPase 13A3 |
| chr5_+_154522119 | 0.01 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr4_+_57715946 | 0.01 |
ENSRNOT00000079059
ENSRNOT00000034429 |
Klhdc10
|
kelch domain containing 10 |
| chr15_+_34270648 | 0.01 |
ENSRNOT00000026333
|
Rnf31
|
ring finger protein 31 |
| chr10_+_84979717 | 0.01 |
ENSRNOT00000065555
|
Mrpl10
|
mitochondrial ribosomal protein L10 |
| chr3_+_155297566 | 0.01 |
ENSRNOT00000021435
ENSRNOT00000084866 |
Dhx35
|
DEAH-box helicase 35 |
| chr7_-_142180794 | 0.01 |
ENSRNOT00000037447
|
Tfcp2
|
transcription factor CP2 |
| chr5_+_14415606 | 0.01 |
ENSRNOT00000089273
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr11_-_4332255 | 0.01 |
ENSRNOT00000087133
|
Cadm2
|
cell adhesion molecule 2 |
| chr1_+_84067938 | 0.01 |
ENSRNOT00000057213
|
Numbl
|
NUMB-like, endocytic adaptor protein |
| chr14_+_84417320 | 0.01 |
ENSRNOT00000008788
|
Tbc1d10a
|
TBC1 domain family, member 10a |
| chr14_-_83219464 | 0.01 |
ENSRNOT00000067694
|
Depdc5
|
DEP domain containing 5 |
| chr1_-_59347472 | 0.01 |
ENSRNOT00000017718
|
Lnpep
|
leucyl and cystinyl aminopeptidase |
| chr18_+_30574627 | 0.01 |
ENSRNOT00000060484
|
Pcdhb19
|
protocadherin beta 19 |
| chr20_+_3827086 | 0.01 |
ENSRNOT00000081588
|
Rxrb
|
retinoid X receptor beta |
| chr1_-_265798167 | 0.01 |
ENSRNOT00000079483
|
Ldb1
|
LIM domain binding 1 |
| chr4_+_113948514 | 0.01 |
ENSRNOT00000011521
|
LOC103692171
|
mannosyl-oligosaccharide glucosidase |
| chr1_+_72425707 | 0.00 |
ENSRNOT00000058956
|
Sbk2
|
SH3 domain binding kinase family, member 2 |
| chr19_-_27535792 | 0.00 |
ENSRNOT00000024040
|
Olr1666
|
olfactory receptor 1666 |
| chr9_+_88494676 | 0.00 |
ENSRNOT00000089451
|
LOC102556337
|
mitochondrial fission factor-like |
| chr6_-_122721496 | 0.00 |
ENSRNOT00000079697
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr2_+_229196616 | 0.00 |
ENSRNOT00000012773
|
Ndst4
|
N-deacetylase and N-sulfotransferase 4 |
| chr5_-_126911520 | 0.00 |
ENSRNOT00000091521
|
Dio1
|
deiodinase, iodothyronine, type I |
| chr16_-_69242028 | 0.00 |
ENSRNOT00000019002
|
Zfp703
|
zinc finger protein 703 |
| chr10_-_102423084 | 0.00 |
ENSRNOT00000065145
|
Sdk2
|
sidekick cell adhesion molecule 2 |
| chrX_-_119162518 | 0.00 |
ENSRNOT00000073176
|
LOC103694506
|
ubiquitin-conjugating enzyme E2 W |
| chr16_+_23553647 | 0.00 |
ENSRNOT00000041994
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr12_+_51878153 | 0.00 |
ENSRNOT00000056798
|
Hscb
|
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr15_-_86142672 | 0.00 |
ENSRNOT00000057830
|
Commd6
|
COMM domain containing 6 |
| chr3_-_13896838 | 0.00 |
ENSRNOT00000025399
|
Fbxw2
|
F-box and WD repeat domain containing 2 |
| chr5_-_76039760 | 0.00 |
ENSRNOT00000019959
|
RGD1306148
|
similar to KIAA0368 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Thra
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0021917 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.0 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.0 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.0 | GO:0036117 | hyaluranon cable(GO:0036117) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |