Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
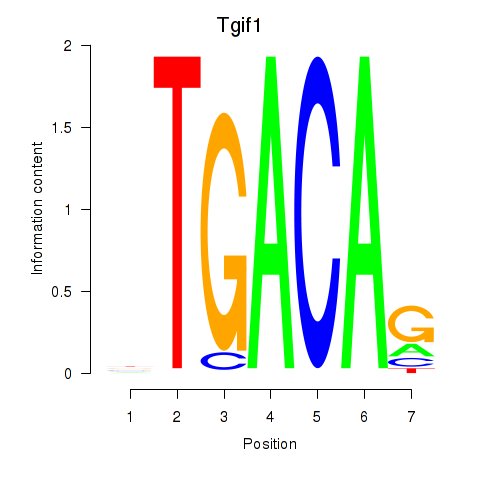
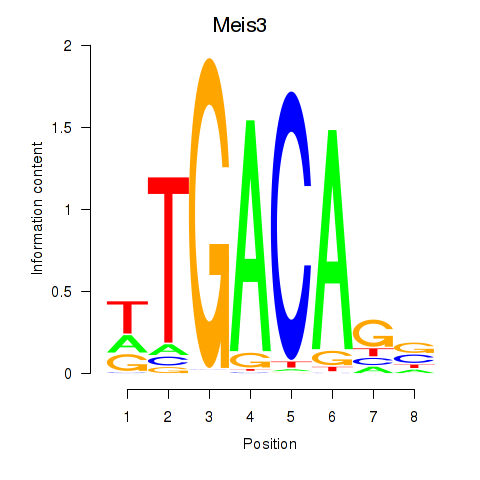
Results for Tgif1_Meis3
Z-value: 1.18
Transcription factors associated with Tgif1_Meis3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tgif1
|
ENSRNOG00000015906 | TGFB-induced factor homeobox 1 |
|
Meis3
|
ENSRNOG00000021390 | Meis homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tgif1 | rn6_v1_chr9_-_119190698_119190698 | 0.70 | 1.9e-01 | Click! |
| Meis3 | rn6_v1_chr1_+_78069193_78069193 | -0.13 | 8.3e-01 | Click! |
Activity profile of Tgif1_Meis3 motif
Sorted Z-values of Tgif1_Meis3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_81819799 | 0.59 |
ENSRNOT00000076840
|
Mxd4
|
Max dimerization protein 4 |
| chr14_+_63095720 | 0.50 |
ENSRNOT00000006071
|
Ppargc1a
|
PPARG coactivator 1 alpha |
| chr15_+_8739589 | 0.50 |
ENSRNOT00000071751
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr13_-_49313940 | 0.48 |
ENSRNOT00000012190
|
Cntn2
|
contactin 2 |
| chr7_-_107774397 | 0.48 |
ENSRNOT00000093322
|
Ndrg1
|
N-myc downstream regulated 1 |
| chr7_-_139907640 | 0.47 |
ENSRNOT00000045473
|
Zfp641
|
zinc finger protein 641 |
| chr2_+_155555840 | 0.44 |
ENSRNOT00000080951
|
AABR07010944.1
|
|
| chr5_-_75319765 | 0.44 |
ENSRNOT00000085698
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr10_-_71358190 | 0.41 |
ENSRNOT00000078646
|
Ac1576
|
uncharacterized LOC102552783 |
| chr11_-_71108184 | 0.40 |
ENSRNOT00000051989
ENSRNOT00000073865 ENSRNOT00000063883 |
Lrch3
|
leucine rich repeats and calponin homology domain containing 3 |
| chr14_-_108658371 | 0.39 |
ENSRNOT00000008919
|
Papolg
|
poly(A) polymerase gamma |
| chr5_-_75319189 | 0.38 |
ENSRNOT00000047200
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr17_-_55709740 | 0.37 |
ENSRNOT00000033359
|
RGD1562037
|
similar to OTTHUMP00000046255 |
| chr7_+_53878610 | 0.37 |
ENSRNOT00000091910
|
Osbpl8
|
oxysterol binding protein-like 8 |
| chr15_+_36865548 | 0.36 |
ENSRNOT00000076460
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr1_+_241594565 | 0.36 |
ENSRNOT00000020123
|
Apba1
|
amyloid beta precursor protein binding family A member 1 |
| chr12_-_35979193 | 0.36 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr2_-_140618405 | 0.35 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr11_+_57207656 | 0.34 |
ENSRNOT00000038207
ENSRNOT00000085754 |
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr5_+_90800082 | 0.33 |
ENSRNOT00000093696
ENSRNOT00000093206 |
Kdm4c
|
lysine demethylase 4C |
| chr4_+_120671489 | 0.33 |
ENSRNOT00000029125
|
Mgll
|
monoglyceride lipase |
| chr10_+_71278650 | 0.32 |
ENSRNOT00000092020
|
Synrg
|
synergin, gamma |
| chr5_+_139963002 | 0.32 |
ENSRNOT00000048506
|
Col9a2
|
collagen type IX alpha 2 chain |
| chr8_+_52729003 | 0.32 |
ENSRNOT00000081738
|
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr11_+_88732381 | 0.32 |
ENSRNOT00000078367
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr4_+_87026530 | 0.31 |
ENSRNOT00000018425
|
Avl9
|
AVL9 cell migration associated |
| chr6_+_60566196 | 0.30 |
ENSRNOT00000006709
ENSRNOT00000075193 |
Dock4
|
dedicator of cytokinesis 4 |
| chr3_+_55094637 | 0.30 |
ENSRNOT00000058763
|
Cers6
|
ceramide synthase 6 |
| chr3_-_112371664 | 0.30 |
ENSRNOT00000079606
ENSRNOT00000012636 |
LOC100910478
|
leucine-rich repeat-containing protein 57-like |
| chrX_-_29648359 | 0.30 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr12_-_10391270 | 0.29 |
ENSRNOT00000092340
|
Wasf3
|
WAS protein family, member 3 |
| chr1_-_277902279 | 0.29 |
ENSRNOT00000078416
|
Ablim1
|
actin-binding LIM protein 1 |
| chr10_+_93811350 | 0.28 |
ENSRNOT00000077280
|
Tanc2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr20_-_27208041 | 0.28 |
ENSRNOT00000084720
|
Rufy2
|
RUN and FYVE domain containing 2 |
| chr17_-_1085885 | 0.28 |
ENSRNOT00000026287
|
Ptch1
|
patched 1 |
| chr11_-_39767802 | 0.28 |
ENSRNOT00000072806
|
RGD1565472
|
similar to RIKEN cDNA 2310003P10 |
| chr8_+_14239594 | 0.28 |
ENSRNOT00000015383
|
Slc36a4
|
solute carrier family 36 member 4 |
| chr9_-_28732919 | 0.27 |
ENSRNOT00000083915
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr3_+_8958090 | 0.27 |
ENSRNOT00000086789
ENSRNOT00000064557 |
Dolpp1
|
dolichyldiphosphatase 1 |
| chr7_-_142210738 | 0.27 |
ENSRNOT00000006095
|
Pou6f1
|
POU class 6 homeobox 1 |
| chr1_+_201981357 | 0.27 |
ENSRNOT00000027999
|
Acadsb
|
acyl-CoA dehydrogenase, short/branched chain |
| chr2_-_198002625 | 0.27 |
ENSRNOT00000091888
|
BC028528
|
cDNA sequence BC028528 |
| chr3_-_125213607 | 0.27 |
ENSRNOT00000078070
|
Gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr14_+_104250617 | 0.27 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr7_+_11033317 | 0.27 |
ENSRNOT00000007498
|
Gna11
|
guanine nucleotide binding protein, alpha 11 |
| chr20_+_44220840 | 0.27 |
ENSRNOT00000047194
|
Tube1
|
tubulin, epsilon 1 |
| chr10_-_36419926 | 0.26 |
ENSRNOT00000004902
|
Znf354b
|
zinc finger protein 354B |
| chr2_+_148262110 | 0.26 |
ENSRNOT00000046641
|
AABR07010705.1
|
|
| chr6_+_64808238 | 0.26 |
ENSRNOT00000093195
|
Nrcam
|
neuronal cell adhesion molecule |
| chr4_+_22445414 | 0.26 |
ENSRNOT00000087657
ENSRNOT00000030224 |
Rundc3b
|
RUN domain containing 3B |
| chr11_-_62451149 | 0.25 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chrX_-_84821775 | 0.25 |
ENSRNOT00000000174
|
Chm
|
CHM, Rab escort protein 1 |
| chr5_+_59080765 | 0.25 |
ENSRNOT00000021888
ENSRNOT00000064419 |
Rgp1
|
RGP1 homolog, RAB6A GEF complex partner 1 |
| chr2_-_187786700 | 0.25 |
ENSRNOT00000092257
ENSRNOT00000092612 ENSRNOT00000068360 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr8_-_116532169 | 0.25 |
ENSRNOT00000085364
|
Rbm5
|
RNA binding motif protein 5 |
| chr13_+_90723092 | 0.25 |
ENSRNOT00000010146
|
Kcnj10
|
ATP-sensitive inward rectifier potassium channel 10 |
| chr3_+_95707386 | 0.25 |
ENSRNOT00000005882
|
Pax6
|
paired box 6 |
| chr8_+_118392773 | 0.24 |
ENSRNOT00000066305
|
LOC100911725
|
6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4-like |
| chr8_-_118696187 | 0.24 |
ENSRNOT00000056141
|
Klhl18
|
kelch-like family member 18 |
| chr16_-_20097287 | 0.24 |
ENSRNOT00000025162
|
Unc13a
|
unc-13 homolog A |
| chr4_-_155740193 | 0.24 |
ENSRNOT00000043229
|
LOC102550396
|
LRRGT00188 |
| chr1_-_128695796 | 0.24 |
ENSRNOT00000076329
|
Synm
|
synemin |
| chr17_+_9596957 | 0.24 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr13_+_97838361 | 0.24 |
ENSRNOT00000003641
|
Cnst
|
consortin, connexin sorting protein |
| chr3_+_11811962 | 0.24 |
ENSRNOT00000060258
ENSRNOT00000039151 |
Ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr18_+_87580424 | 0.24 |
ENSRNOT00000070984
|
Tmx3
|
thioredoxin-related transmembrane protein 3 |
| chr1_-_265573117 | 0.24 |
ENSRNOT00000044195
ENSRNOT00000055915 |
LOC100911951
|
Kv channel-interacting protein 2-like |
| chr14_-_81254637 | 0.23 |
ENSRNOT00000058166
|
Htt
|
huntingtin |
| chr4_-_59809321 | 0.23 |
ENSRNOT00000017536
|
Plxna4
|
plexin A4 |
| chr7_+_20262680 | 0.23 |
ENSRNOT00000046378
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr4_-_129515435 | 0.23 |
ENSRNOT00000039353
|
Eogt
|
EGF domain specific O-linked N-acetylglucosamine transferase |
| chr18_+_15856801 | 0.23 |
ENSRNOT00000071548
|
Zfp397
|
zinc finger protein 397 |
| chr1_-_103128743 | 0.23 |
ENSRNOT00000075006
|
Spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr11_+_89511191 | 0.23 |
ENSRNOT00000002510
|
Mcm4
|
minichromosome maintenance complex component 4 |
| chr1_+_247519939 | 0.22 |
ENSRNOT00000034421
|
Cd274
|
CD274 molecule |
| chr5_-_59647633 | 0.22 |
ENSRNOT00000091944
|
Rnf38
|
ring finger protein 38 |
| chr18_+_74299931 | 0.22 |
ENSRNOT00000078403
|
Epg5
|
ectopic P-granules autophagy protein 5 homolog |
| chr16_-_19791832 | 0.22 |
ENSRNOT00000040393
|
Ushbp1
|
USH1 protein network component harmonin binding protein 1 |
| chr8_-_49280901 | 0.22 |
ENSRNOT00000021390
|
Cd3g
|
CD3g molecule |
| chr10_-_5196892 | 0.22 |
ENSRNOT00000083982
|
Clec16a
|
C-type lectin domain family 16, member A |
| chr17_+_88215834 | 0.22 |
ENSRNOT00000034098
|
Gpr158
|
G protein-coupled receptor 158 |
| chr1_-_259484569 | 0.22 |
ENSRNOT00000021289
ENSRNOT00000021114 ENSRNOT00000021229 ENSRNOT00000021347 ENSRNOT00000021226 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr11_+_71151132 | 0.21 |
ENSRNOT00000082594
ENSRNOT00000082435 |
Rubcn
|
RUN and cysteine rich domain containing beclin 1 interacting protein |
| chr1_-_82344345 | 0.21 |
ENSRNOT00000051892
ENSRNOT00000090629 ENSRNOT00000029487 ENSRNOT00000027933 |
Ceacam1
|
carcinoembryonic antigen related cell adhesion molecule 1 |
| chr10_+_75055020 | 0.21 |
ENSRNOT00000010755
|
Tspoap1
|
TSPO associated protein 1 |
| chr3_+_134413170 | 0.21 |
ENSRNOT00000074338
|
AABR07054000.1
|
|
| chr1_-_65681440 | 0.21 |
ENSRNOT00000026305
|
Zfp128
|
zinc finger protein 128 |
| chr6_+_10348308 | 0.21 |
ENSRNOT00000034991
|
Epas1
|
endothelial PAS domain protein 1 |
| chr7_-_120874308 | 0.21 |
ENSRNOT00000078320
|
Fam227a
|
family with sequence similarity 227, member A |
| chr20_+_44803885 | 0.21 |
ENSRNOT00000077936
|
Rev3l
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr7_+_27174882 | 0.21 |
ENSRNOT00000051181
|
Glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr2_+_187512164 | 0.21 |
ENSRNOT00000051394
|
Mef2d
|
myocyte enhancer factor 2D |
| chr3_-_111354419 | 0.21 |
ENSRNOT00000038365
|
Exd1
|
exonuclease 3'-5' domain containing 1 |
| chr1_-_166912524 | 0.21 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr1_-_219412816 | 0.21 |
ENSRNOT00000083204
ENSRNOT00000029580 |
Rps6kb2
|
ribosomal protein S6 kinase B2 |
| chr3_-_71845232 | 0.21 |
ENSRNOT00000078645
|
Calcrl
|
calcitonin receptor like receptor |
| chr3_+_176821652 | 0.21 |
ENSRNOT00000055030
|
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr20_-_6251634 | 0.21 |
ENSRNOT00000059194
|
Stk38
|
serine/threonine kinase 38 |
| chr1_+_264670841 | 0.20 |
ENSRNOT00000034814
ENSRNOT00000082412 |
Slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr1_-_37886675 | 0.20 |
ENSRNOT00000024149
|
RGD1566325
|
similar to regulator of sex-limitation candidate 16 |
| chr6_-_115616766 | 0.20 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chr12_-_29743705 | 0.20 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr14_+_9226125 | 0.20 |
ENSRNOT00000088047
|
Wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr4_+_88834066 | 0.20 |
ENSRNOT00000009546
|
Abcg2
|
ATP-binding cassette, subfamily G (WHITE), member 2 |
| chr1_+_221673590 | 0.20 |
ENSRNOT00000038016
|
Cdc42bpg
|
CDC42 binding protein kinase gamma |
| chr6_+_58468155 | 0.20 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr10_-_95407997 | 0.20 |
ENSRNOT00000077888
|
Nol11
|
nucleolar protein 11 |
| chr3_-_120157866 | 0.20 |
ENSRNOT00000020367
|
LOC100909998
|
zinc finger protein 2-like |
| chr6_+_64224861 | 0.20 |
ENSRNOT00000093159
ENSRNOT00000093664 |
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr1_-_66237501 | 0.20 |
ENSRNOT00000073006
|
Zfp606
|
zinc finger protein 606 |
| chr8_-_47339343 | 0.20 |
ENSRNOT00000081007
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr10_-_16045835 | 0.20 |
ENSRNOT00000064832
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr10_+_103395511 | 0.20 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr5_+_4373626 | 0.19 |
ENSRNOT00000064946
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr10_-_46720907 | 0.19 |
ENSRNOT00000083093
ENSRNOT00000067866 |
Tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr10_+_106712127 | 0.19 |
ENSRNOT00000040629
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr14_-_77810147 | 0.19 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chr16_+_23668595 | 0.19 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_+_27476375 | 0.19 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr17_-_67904674 | 0.19 |
ENSRNOT00000078532
|
Klf6
|
Kruppel-like factor 6 |
| chr1_-_65611815 | 0.19 |
ENSRNOT00000036874
|
Zfp324
|
zinc finger protein 324 |
| chr10_-_49149479 | 0.19 |
ENSRNOT00000004291
|
Zfp287
|
zinc finger protein 287 |
| chr10_-_15306515 | 0.19 |
ENSRNOT00000060150
|
Nhlrc4
|
NHL repeat containing 4 |
| chr9_+_94425252 | 0.19 |
ENSRNOT00000064965
ENSRNOT00000076099 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr12_+_660011 | 0.19 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr19_-_57649827 | 0.19 |
ENSRNOT00000026757
|
Exoc8
|
exocyst complex component 8 |
| chr1_+_87563975 | 0.19 |
ENSRNOT00000088772
|
Zfp30
|
zinc finger protein 30 |
| chr8_+_29453643 | 0.18 |
ENSRNOT00000090643
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr18_+_6207280 | 0.18 |
ENSRNOT00000037955
|
Taf4b
|
TATA-box binding protein associated factor 4b |
| chr13_+_91054974 | 0.18 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr6_-_108796124 | 0.18 |
ENSRNOT00000086545
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr13_+_44475970 | 0.18 |
ENSRNOT00000024602
ENSRNOT00000091645 |
Ccnt2
|
cyclin T2 |
| chr10_+_93811505 | 0.18 |
ENSRNOT00000081937
|
Tanc2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr13_-_48848864 | 0.18 |
ENSRNOT00000077857
ENSRNOT00000068003 |
Mfsd4
|
major facilitator superfamily domain containing 4 |
| chr1_-_235405831 | 0.18 |
ENSRNOT00000071578
|
AABR07006458.1
|
|
| chr1_-_66212418 | 0.18 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr2_+_219628695 | 0.18 |
ENSRNOT00000067324
|
Sass6
|
SAS-6 centriolar assembly protein |
| chr18_+_38847632 | 0.18 |
ENSRNOT00000014916
|
LOC102554034
|
ELMO domain-containing protein 2-like |
| chr5_-_169212170 | 0.18 |
ENSRNOT00000013385
|
Tas1r1
|
taste 1 receptor member 1 |
| chr11_+_33929163 | 0.18 |
ENSRNOT00000083594
|
Dopey2
|
dopey family member 2 |
| chr3_-_80012750 | 0.17 |
ENSRNOT00000018154
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr1_+_61522298 | 0.17 |
ENSRNOT00000029111
|
Zfp51
|
zinc finger protein 51 |
| chr11_-_68197772 | 0.17 |
ENSRNOT00000003060
ENSRNOT00000081875 |
Hspbap1
|
Hspb associated protein 1 |
| chr19_+_30668602 | 0.17 |
ENSRNOT00000035084
|
Usp38
|
ubiquitin specific peptidase 38 |
| chr7_-_120967490 | 0.17 |
ENSRNOT00000046399
|
Sun2
|
Sad1 and UNC84 domain containing 2 |
| chr18_-_55992885 | 0.17 |
ENSRNOT00000025881
|
Ndst1
|
N-deacetylase and N-sulfotransferase 1 |
| chr11_-_71591502 | 0.17 |
ENSRNOT00000002403
|
Pcyt1a
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr17_-_86657473 | 0.17 |
ENSRNOT00000078827
|
AABR07028795.1
|
|
| chr4_-_22424862 | 0.17 |
ENSRNOT00000082359
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr7_-_115045802 | 0.17 |
ENSRNOT00000040236
ENSRNOT00000071818 |
Mroh5
|
maestro heat-like repeat family member 5 |
| chr3_+_151126591 | 0.17 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr19_-_19832812 | 0.17 |
ENSRNOT00000084657
|
Papd5
|
poly(A) RNA polymerase D5, non-canonical |
| chr4_-_177331874 | 0.17 |
ENSRNOT00000065387
ENSRNOT00000091099 |
C2cd5
|
C2 calcium-dependent domain containing 5 |
| chr7_+_130498199 | 0.17 |
ENSRNOT00000092684
ENSRNOT00000092431 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr15_+_39638510 | 0.17 |
ENSRNOT00000037800
|
Rcbtb1
|
RCC1 and BTB domain containing protein 1 |
| chr1_+_167066516 | 0.17 |
ENSRNOT00000000474
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr6_+_54438436 | 0.17 |
ENSRNOT00000091972
|
AABR07063893.1
|
|
| chr9_+_53627208 | 0.17 |
ENSRNOT00000083487
|
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr12_-_18487178 | 0.17 |
ENSRNOT00000030757
|
Zfp157
|
zinc finger protein 157 |
| chr3_+_152571121 | 0.17 |
ENSRNOT00000087289
ENSRNOT00000080543 ENSRNOT00000083476 |
Epb41l1
|
erythrocyte membrane protein band 4.1-like 1 |
| chrX_-_54303729 | 0.17 |
ENSRNOT00000087919
ENSRNOT00000064340 ENSRNOT00000051249 ENSRNOT00000087547 |
Gk
|
glycerol kinase |
| chr19_+_43882249 | 0.17 |
ENSRNOT00000025761
|
Zfp1
|
zinc finger protein 1 |
| chr2_+_198231291 | 0.17 |
ENSRNOT00000078288
ENSRNOT00000046739 |
Otud7b
|
OTU deubiquitinase 7B |
| chr13_+_50196042 | 0.17 |
ENSRNOT00000090858
|
Zc3h11a
|
zinc finger CCCH-type containing 11A |
| chr3_-_161376119 | 0.17 |
ENSRNOT00000023521
|
Zfp335
|
zinc finger protein 335 |
| chr4_+_157594436 | 0.17 |
ENSRNOT00000029053
|
Lpar5
|
lysophosphatidic acid receptor 5 |
| chr7_+_66717267 | 0.17 |
ENSRNOT00000005654
|
Mon2
|
MON2 homolog, regulator of endosome-to-Golgi trafficking |
| chr5_+_5616483 | 0.17 |
ENSRNOT00000011026
|
Ncoa2
|
nuclear receptor coactivator 2 |
| chr10_-_37215899 | 0.17 |
ENSRNOT00000006356
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr3_-_66279155 | 0.17 |
ENSRNOT00000079887
|
Cerkl
|
ceramide kinase-like |
| chr18_+_14756684 | 0.17 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr14_-_114692764 | 0.17 |
ENSRNOT00000008210
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr10_-_37209881 | 0.17 |
ENSRNOT00000090475
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr7_+_13318533 | 0.16 |
ENSRNOT00000034545
|
LOC691170
|
similar to zinc finger protein 84 (HPF2) |
| chrX_-_10031167 | 0.16 |
ENSRNOT00000060988
|
Gpr34
|
G protein-coupled receptor 34 |
| chr1_+_252944103 | 0.16 |
ENSRNOT00000025770
|
Ifit1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr2_-_80667481 | 0.16 |
ENSRNOT00000016784
|
Trio
|
trio Rho guanine nucleotide exchange factor |
| chr12_-_21362205 | 0.16 |
ENSRNOT00000064787
|
LOC108348155
|
paired immunoglobulin-like type 2 receptor beta-2 |
| chr2_+_242882306 | 0.16 |
ENSRNOT00000013661
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr12_-_2555164 | 0.16 |
ENSRNOT00000084460
ENSRNOT00000061821 |
Map2k7
|
mitogen activated protein kinase kinase 7 |
| chr2_+_72006099 | 0.16 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr13_-_100740728 | 0.16 |
ENSRNOT00000000074
|
Fbxo28
|
F-box protein 28 |
| chr15_-_77736892 | 0.16 |
ENSRNOT00000057924
|
Pcdh9
|
protocadherin 9 |
| chr4_+_168599331 | 0.16 |
ENSRNOT00000086719
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr2_+_86891092 | 0.16 |
ENSRNOT00000077162
ENSRNOT00000083066 |
LOC100360380
|
zinc finger protein 457-like |
| chr7_-_26361221 | 0.16 |
ENSRNOT00000011326
|
RGD1309995
|
similar to CG13957-PA |
| chr6_-_72596446 | 0.16 |
ENSRNOT00000008459
|
Hectd1
|
HECT domain E3 ubiquitin protein ligase 1 |
| chr4_+_181315444 | 0.16 |
ENSRNOT00000044147
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr7_+_2752680 | 0.16 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr19_+_755460 | 0.16 |
ENSRNOT00000076560
|
Dync1li2
|
dynein, cytoplasmic 1 light intermediate chain 2 |
| chr5_+_126226291 | 0.16 |
ENSRNOT00000009582
|
Ttc22
|
tetratricopeptide repeat domain 22 |
| chr7_-_64887448 | 0.16 |
ENSRNOT00000038147
|
Helb
|
DNA helicase B |
| chr7_-_82687130 | 0.16 |
ENSRNOT00000006791
|
Tmem74
|
transmembrane protein 74 |
| chr9_-_119818310 | 0.16 |
ENSRNOT00000019359
|
Smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr12_-_20276121 | 0.16 |
ENSRNOT00000065873
|
LOC685157
|
similar to paired immunoglobin-like type 2 receptor beta |
| chr7_-_120882392 | 0.16 |
ENSRNOT00000056179
ENSRNOT00000056178 |
Fam227a
|
family with sequence similarity 227, member A |
| chr20_+_14101659 | 0.15 |
ENSRNOT00000072696
|
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr16_-_37177033 | 0.15 |
ENSRNOT00000014015
|
Fbxo8
|
F-box protein 8 |
| chr4_-_118472179 | 0.15 |
ENSRNOT00000023856
|
Mxd1
|
max dimerization protein 1 |
| chr5_-_79899054 | 0.15 |
ENSRNOT00000074379
|
AC229945.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Tgif1_Meis3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1904638 | response to resveratrol(GO:1904638) |
| 0.2 | 0.5 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.4 | GO:0003322 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.1 | 0.3 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.4 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.3 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.3 | GO:0030473 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.3 | GO:0015827 | tryptophan transport(GO:0015827) |
| 0.1 | 0.4 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 0.3 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 0.1 | 0.4 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.2 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 0.3 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.2 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.1 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.2 | GO:0070237 | negative regulation of T cell mediated cytotoxicity(GO:0001915) positive regulation of activation-induced cell death of T cells(GO:0070237) insulin catabolic process(GO:1901143) |
| 0.1 | 0.2 | GO:1903179 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.1 | 0.3 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 0.2 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.2 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.1 | 0.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 | 0.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.4 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 0.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.2 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.1 | 0.2 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.2 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:1902962 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.2 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.1 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.5 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.2 | GO:1990963 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 0.0 | 0.2 | GO:1904008 | response to monosodium glutamate(GO:1904008) |
| 0.0 | 0.2 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.3 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.3 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.2 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) regulation of cellular amino acid biosynthetic process(GO:2000282) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.4 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.1 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.0 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.0 | 0.2 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.1 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:2000458 | astrocyte chemotaxis(GO:0035700) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.2 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.5 | GO:0099562 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.3 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:1905065 | positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.1 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.2 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0072275 | cell migration involved in vasculogenesis(GO:0035441) VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:1903373 | positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) positive regulation of oocyte maturation(GO:1900195) |
| 0.0 | 0.1 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.0 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.0 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.0 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.1 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 0.1 | GO:0042357 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.0 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.1 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.1 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.4 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.2 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.0 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.0 | 0.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.0 | 0.1 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.2 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0001582 | detection of chemical stimulus involved in sensory perception of sweet taste(GO:0001582) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0098907 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.0 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) tarsal gland development(GO:1903699) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.2 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.0 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.0 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.1 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.2 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0043697 | thelarche(GO:0042695) dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.0 | GO:0070627 | ferrous iron import(GO:0070627) |
| 0.0 | 0.1 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.0 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.1 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0019471 | 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.3 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.2 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.1 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0070427 | positive regulation of immature T cell proliferation(GO:0033091) positive regulation of T-helper 1 cell differentiation(GO:0045627) nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.0 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.0 | 0.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.3 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.0 | GO:2000407 | regulation of T cell extravasation(GO:2000407) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.0 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.0 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.0 | 0.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.0 | GO:0070537 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.0 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.0 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.0 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.0 | GO:0035600 | tRNA methylthiolation(GO:0035600) |
| 0.0 | 0.0 | GO:0051938 | L-glutamate import(GO:0051938) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.0 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.0 | GO:1903874 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.0 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.0 | 0.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.0 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.0 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.0 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.7 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.2 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0090534 | calcium ion-transporting ATPase complex(GO:0090534) |
| 0.0 | 0.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0099571 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.0 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.0 | 0.1 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:1903767 | sweet taste receptor complex(GO:1903767) taste receptor complex(GO:1903768) |
| 0.0 | 0.0 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.0 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.3 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.2 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.1 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.2 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.3 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) |
| 0.0 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.5 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.0 | 0.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0051185 | adenosine-diphosphatase activity(GO:0043262) coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0008458 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.0 | GO:0001635 | calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.1 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.0 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.0 | GO:0031770 | growth hormone-releasing hormone receptor binding(GO:0031770) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.0 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.0 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.0 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.0 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |