Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
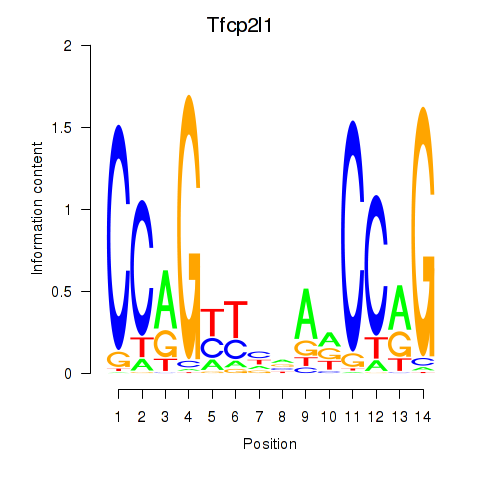
Results for Tfcp2l1
Z-value: 0.33
Transcription factors associated with Tfcp2l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfcp2l1
|
ENSRNOG00000002414 | transcription factor CP2-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfcp2l1 | rn6_v1_chr13_+_34610684_34610689 | -0.42 | 4.8e-01 | Click! |
Activity profile of Tfcp2l1 motif
Sorted Z-values of Tfcp2l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_195908037 | 0.12 |
ENSRNOT00000079776
|
Tuft1
|
tuftelin 1 |
| chr5_-_166726794 | 0.10 |
ENSRNOT00000022799
|
Slc25a33
|
solute carrier family 25 member 33 |
| chr5_-_153924896 | 0.10 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr6_+_86131242 | 0.10 |
ENSRNOT00000039337
|
Fau
|
FAU, ubiquitin like and ribosomal protein S30 fusion |
| chr13_+_104284660 | 0.10 |
ENSRNOT00000005400
|
Dusp10
|
dual specificity phosphatase 10 |
| chr20_+_5049496 | 0.10 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr1_-_141481315 | 0.09 |
ENSRNOT00000020229
|
Pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr11_-_36479868 | 0.08 |
ENSRNOT00000075762
|
LOC100911295
|
non-histone chromosomal protein HMG-14-like |
| chr10_-_97756521 | 0.07 |
ENSRNOT00000045902
|
Slc16a6
|
solute carrier family 16, member 6 |
| chr3_+_176691329 | 0.07 |
ENSRNOT00000017319
|
Ppdpf
|
pancreatic progenitor cell differentiation and proliferation factor |
| chr6_+_42852683 | 0.07 |
ENSRNOT00000079185
|
Odc1
|
ornithine decarboxylase 1 |
| chr9_+_81672758 | 0.06 |
ENSRNOT00000020646
|
Ctdsp1
|
CTD small phosphatase 1 |
| chr1_-_164205222 | 0.06 |
ENSRNOT00000035774
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr1_+_147021436 | 0.06 |
ENSRNOT00000042490
|
F8a1
|
coagulation factor VIII-associated 1 |
| chr1_+_264504591 | 0.06 |
ENSRNOT00000050076
|
Pax2
|
paired box 2 |
| chr13_+_47454591 | 0.06 |
ENSRNOT00000005791
|
LOC498222
|
similar to specifically androgen-regulated protein |
| chr10_-_105116916 | 0.06 |
ENSRNOT00000012373
|
Evpl
|
envoplakin |
| chr7_+_79138925 | 0.06 |
ENSRNOT00000047511
|
AABR07057586.1
|
|
| chr5_-_164747083 | 0.06 |
ENSRNOT00000010433
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr6_+_23757225 | 0.06 |
ENSRNOT00000005553
|
LOC298795
|
similar to 14-3-3 protein sigma |
| chr5_-_152198813 | 0.05 |
ENSRNOT00000082953
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr1_-_225128740 | 0.05 |
ENSRNOT00000026897
|
Rom1
|
retinal outer segment membrane protein 1 |
| chr1_-_242373764 | 0.05 |
ENSRNOT00000076544
|
Fam122a
|
family with sequence similarity 122A |
| chr17_+_30965942 | 0.05 |
ENSRNOT00000060143
|
Pxdc1
|
PX domain containing 1 |
| chr2_-_204254699 | 0.05 |
ENSRNOT00000021487
|
Mab21l3
|
mab-21 like 3 |
| chr2_+_189400696 | 0.05 |
ENSRNOT00000046919
ENSRNOT00000089801 |
LOC361985
|
similar to NICE-3 |
| chr16_-_36080191 | 0.04 |
ENSRNOT00000017635
|
Hmgb2l1
|
high mobility group box 2-like 1 |
| chr14_-_91904433 | 0.04 |
ENSRNOT00000005857
|
Fignl1
|
fidgetin-like 1 |
| chr8_-_50231357 | 0.04 |
ENSRNOT00000081737
|
Tagln
|
transgelin |
| chr2_+_189106039 | 0.04 |
ENSRNOT00000028210
|
Ube2q1
|
ubiquitin conjugating enzyme E2 Q1 |
| chr3_+_113319456 | 0.04 |
ENSRNOT00000051354
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr8_+_71719563 | 0.04 |
ENSRNOT00000022828
|
Ppib
|
peptidylprolyl isomerase B |
| chr7_-_139483997 | 0.04 |
ENSRNOT00000086062
|
Col2a1
|
collagen type II alpha 1 chain |
| chr8_+_116754178 | 0.04 |
ENSRNOT00000068295
ENSRNOT00000084429 |
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr10_+_66509107 | 0.04 |
ENSRNOT00000017230
|
RGD1565317
|
similar to ubiquitin-like/S30 ribosomal fusion protein |
| chr9_+_81518176 | 0.04 |
ENSRNOT00000078317
ENSRNOT00000019265 ENSRNOT00000088246 ENSRNOT00000084682 |
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr13_+_52854031 | 0.04 |
ENSRNOT00000013738
ENSRNOT00000086004 |
Tmem9
|
transmembrane protein 9 |
| chr4_+_145427367 | 0.04 |
ENSRNOT00000037788
|
Il17rc
|
interleukin 17 receptor C |
| chrX_-_111887906 | 0.03 |
ENSRNOT00000085118
|
Tsc22d3
|
TSC22 domain family, member 3 |
| chr11_+_39482408 | 0.03 |
ENSRNOT00000075126
|
Hmgn1
|
high mobility group nucleosome binding domain 1 |
| chr12_+_15890170 | 0.03 |
ENSRNOT00000001654
|
Gna12
|
G protein subunit alpha 12 |
| chr13_-_49487892 | 0.03 |
ENSRNOT00000044972
|
Nfasc
|
neurofascin |
| chr20_+_3553455 | 0.03 |
ENSRNOT00000090080
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr7_+_98770840 | 0.03 |
ENSRNOT00000011891
|
Rnf139
|
ring finger protein 139 |
| chr9_-_17114177 | 0.03 |
ENSRNOT00000061185
|
Lrrc73
|
leucine rich repeat containing 73 |
| chr4_+_21317695 | 0.03 |
ENSRNOT00000007572
|
Grm3
|
glutamate metabotropic receptor 3 |
| chr20_-_11620945 | 0.03 |
ENSRNOT00000079725
|
Krtap12-2
|
keratin associated protein 12-2 |
| chr8_+_117679278 | 0.02 |
ENSRNOT00000042114
|
Uqcrc1
|
ubiquinol-cytochrome c reductase core protein I |
| chr10_+_55675575 | 0.02 |
ENSRNOT00000057295
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr5_-_14356692 | 0.02 |
ENSRNOT00000085654
|
Atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr8_+_115151627 | 0.02 |
ENSRNOT00000064252
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr7_-_116936674 | 0.02 |
ENSRNOT00000029456
|
Eef1d
|
eukaryotic translation elongation factor 1 delta |
| chr11_+_36075709 | 0.02 |
ENSRNOT00000002247
|
Ets2
|
ETS proto-oncogene 2, transcription factor |
| chr3_+_161236898 | 0.02 |
ENSRNOT00000082303
ENSRNOT00000020323 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr12_-_19582185 | 0.02 |
ENSRNOT00000060020
ENSRNOT00000085853 |
RGD1305455
|
similar to hypothetical protein FLJ10925 |
| chr1_-_199037267 | 0.02 |
ENSRNOT00000078779
|
Ccdc189
|
coiled-coil domain containing 189 |
| chr9_+_81518584 | 0.02 |
ENSRNOT00000084309
|
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr13_-_104284068 | 0.02 |
ENSRNOT00000005407
|
LOC100912365
|
uncharacterized LOC100912365 |
| chr8_-_6034142 | 0.02 |
ENSRNOT00000014560
|
Birc2
|
baculoviral IAP repeat-containing 2 |
| chr3_+_11476883 | 0.02 |
ENSRNOT00000072241
|
Naif1
|
nuclear apoptosis inducing factor 1 |
| chr19_+_56179111 | 0.02 |
ENSRNOT00000046155
|
Tcf25
|
transcription factor 25 |
| chr11_-_28527890 | 0.02 |
ENSRNOT00000002138
|
Cldn8
|
claudin 8 |
| chr2_+_58462588 | 0.02 |
ENSRNOT00000083799
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr5_-_58124681 | 0.02 |
ENSRNOT00000019795
|
Sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr4_-_116278615 | 0.01 |
ENSRNOT00000020505
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr4_+_130332076 | 0.01 |
ENSRNOT00000083127
|
Mitf
|
melanogenesis associated transcription factor |
| chr19_+_32188267 | 0.01 |
ENSRNOT00000025079
|
Smad1
|
SMAD family member 1 |
| chr10_+_61403130 | 0.01 |
ENSRNOT00000092490
ENSRNOT00000077649 |
Ccdc92b
|
coiled-coil domain containing 92B |
| chr7_-_143228060 | 0.01 |
ENSRNOT00000088923
ENSRNOT00000012640 |
Krt75
|
keratin 75 |
| chr1_+_44446765 | 0.01 |
ENSRNOT00000030132
|
Cldn20
|
claudin 20 |
| chr20_+_5080070 | 0.01 |
ENSRNOT00000086950
ENSRNOT00000077082 |
Abhd16a
|
abhydrolase domain containing 16A |
| chr8_+_113603533 | 0.01 |
ENSRNOT00000017280
|
Mrpl3
|
mitochondrial ribosomal protein L3 |
| chr1_+_228690119 | 0.01 |
ENSRNOT00000087169
|
Pfpl
|
pore forming protein-like |
| chr2_+_199199845 | 0.01 |
ENSRNOT00000020932
|
LOC100909441
|
tubulin alpha-1C chain-like |
| chr8_-_59239954 | 0.01 |
ENSRNOT00000016104
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr13_-_82006005 | 0.01 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr3_-_8659102 | 0.01 |
ENSRNOT00000050908
|
Zdhhc12
|
zinc finger, DHHC-type containing 12 |
| chr14_+_108007724 | 0.01 |
ENSRNOT00000014062
|
Xpo1
|
exportin 1 |
| chrX_+_106253355 | 0.00 |
ENSRNOT00000004252
|
LOC102552920
|
armadillo repeat-containing X-linked protein 5-like |
| chr1_+_225129097 | 0.00 |
ENSRNOT00000026938
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr8_-_21661259 | 0.00 |
ENSRNOT00000068308
|
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chr8_-_117446019 | 0.00 |
ENSRNOT00000042577
|
Arih2
|
ariadne RBR E3 ubiquitin protein ligase 2 |
| chr15_+_49069145 | 0.00 |
ENSRNOT00000019487
|
Scara5
|
scavenger receptor class A, member 5 |
| chr14_-_83685273 | 0.00 |
ENSRNOT00000026134
|
Rnf185
|
ring finger protein 185 |
| chr10_+_59893067 | 0.00 |
ENSRNOT00000056443
|
Spata22
|
spermatogenesis associated 22 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfcp2l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:2000814 | positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.0 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.0 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |