Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
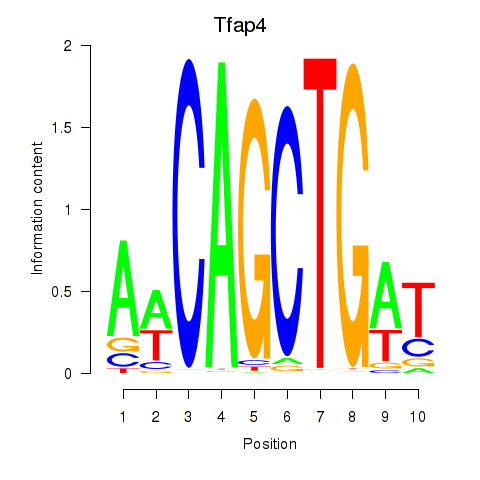
Results for Tfap4
Z-value: 0.64
Transcription factors associated with Tfap4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap4
|
ENSRNOG00000005227 | transcription factor AP-4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap4 | rn6_v1_chr10_+_11206226_11206226 | 0.41 | 4.9e-01 | Click! |
Activity profile of Tfap4 motif
Sorted Z-values of Tfap4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_157538303 | 0.59 |
ENSRNOT00000086418
|
Zfp384
|
zinc finger protein 384 |
| chr10_+_85301875 | 0.41 |
ENSRNOT00000080935
|
Socs7
|
suppressor of cytokine signaling 7 |
| chr17_+_12762752 | 0.36 |
ENSRNOT00000044842
|
Diras2
|
DIRAS family GTPase 2 |
| chr5_+_90800082 | 0.36 |
ENSRNOT00000093696
ENSRNOT00000093206 |
Kdm4c
|
lysine demethylase 4C |
| chr2_-_140618405 | 0.34 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr9_-_66033871 | 0.30 |
ENSRNOT00000035209
|
Als2
|
ALS2, alsin Rho guanine nucleotide exchange factor |
| chr1_-_219745654 | 0.23 |
ENSRNOT00000054848
|
LOC689065
|
hypothetical protein LOC689065 |
| chr9_+_94425252 | 0.22 |
ENSRNOT00000064965
ENSRNOT00000076099 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr10_-_64642292 | 0.22 |
ENSRNOT00000084670
|
Abr
|
active BCR-related |
| chrX_+_62754634 | 0.21 |
ENSRNOT00000016669
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta |
| chr14_+_37113210 | 0.20 |
ENSRNOT00000089094
|
Sgcb
|
sarcoglycan, beta |
| chr8_-_112807598 | 0.19 |
ENSRNOT00000015344
|
Dnajc13
|
DnaJ heat shock protein family (Hsp40) member C13 |
| chr1_+_213583606 | 0.19 |
ENSRNOT00000088899
|
AC109844.1
|
|
| chr1_-_277768368 | 0.18 |
ENSRNOT00000023113
|
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr4_+_199916 | 0.18 |
ENSRNOT00000009317
|
Htr5a
|
5-hydroxytryptamine receptor 5A |
| chr3_-_61494778 | 0.17 |
ENSRNOT00000068018
|
Lnpk
|
lunapark, ER junction formation factor |
| chr5_-_58484900 | 0.17 |
ENSRNOT00000012386
|
Fam214b
|
family with sequence similarity 214, member B |
| chr3_-_148932878 | 0.17 |
ENSRNOT00000013881
|
Nol4l
|
nucleolar protein 4-like |
| chr1_-_215846911 | 0.16 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr10_-_62814520 | 0.15 |
ENSRNOT00000019410
|
Ssh2
|
slingshot protein phosphatase 2 |
| chr7_+_73588163 | 0.15 |
ENSRNOT00000015707
|
Kcns2
|
potassium voltage-gated channel, modifier subfamily S, member 2 |
| chr2_+_114386019 | 0.15 |
ENSRNOT00000082148
|
AABR07009834.1
|
|
| chr10_-_62287189 | 0.15 |
ENSRNOT00000004365
|
Wdr81
|
WD repeat domain 81 |
| chr9_+_111028824 | 0.15 |
ENSRNOT00000041418
ENSRNOT00000056457 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_-_7422738 | 0.14 |
ENSRNOT00000088339
|
Gtf3c4
|
general transcription factor IIIC subunit 4 |
| chrX_+_15610230 | 0.14 |
ENSRNOT00000012880
|
Ccdc120
|
coiled-coil domain containing 120 |
| chr4_-_159482869 | 0.13 |
ENSRNOT00000088333
|
Rad51ap1
|
RAD51 associated protein 1 |
| chr5_-_137372524 | 0.13 |
ENSRNOT00000009061
|
Tmem125
|
transmembrane protein 125 |
| chr8_+_70884491 | 0.13 |
ENSRNOT00000072939
|
Ubap1l
|
ubiquitin associated protein 1-like |
| chr1_+_13595295 | 0.13 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr19_-_41433346 | 0.13 |
ENSRNOT00000022952
|
Cmtr2
|
cap methyltransferase 2 |
| chr11_-_24641820 | 0.13 |
ENSRNOT00000044081
ENSRNOT00000048854 |
App
|
amyloid beta precursor protein |
| chr9_-_85528860 | 0.13 |
ENSRNOT00000063834
ENSRNOT00000088963 |
Wdfy1
|
WD repeat and FYVE domain containing 1 |
| chr4_+_179481263 | 0.13 |
ENSRNOT00000021284
|
Etfrf1
|
electron transfer flavoprotein regulatory factor 1 |
| chr4_-_157358262 | 0.13 |
ENSRNOT00000021481
|
Gnb3
|
G protein subunit beta 3 |
| chr17_-_10208360 | 0.13 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr4_-_482645 | 0.12 |
ENSRNOT00000062073
ENSRNOT00000071713 |
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr16_+_32457521 | 0.12 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr11_-_90406797 | 0.12 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
| chr20_+_20236151 | 0.12 |
ENSRNOT00000079630
|
Ank3
|
ankyrin 3 |
| chr10_+_70298519 | 0.12 |
ENSRNOT00000035730
ENSRNOT00000076233 ENSRNOT00000076445 ENSRNOT00000076133 |
Slfn5
|
schlafen family member 5 |
| chr13_-_53224768 | 0.12 |
ENSRNOT00000091015
|
Camsap2
|
calmodulin regulated spectrin-associated protein family, member 2 |
| chr12_-_25638797 | 0.11 |
ENSRNOT00000002033
|
Gatsl2
|
GATS protein-like 2 |
| chr7_-_145450233 | 0.11 |
ENSRNOT00000092974
ENSRNOT00000021523 |
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr7_-_107616038 | 0.11 |
ENSRNOT00000088752
|
Sla
|
src-like adaptor |
| chr18_+_27424328 | 0.11 |
ENSRNOT00000033784
|
Kif20a
|
kinesin family member 20A |
| chr8_+_68526093 | 0.11 |
ENSRNOT00000011385
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chr3_-_151625644 | 0.11 |
ENSRNOT00000026657
|
Rbm12
|
RNA binding motif protein 12 |
| chr8_-_50126413 | 0.11 |
ENSRNOT00000072712
ENSRNOT00000022583 |
Cep164
|
centrosomal protein 164 |
| chr1_-_281756159 | 0.11 |
ENSRNOT00000013170
|
Prlhr
|
prolactin releasing hormone receptor |
| chr1_+_15642153 | 0.11 |
ENSRNOT00000079845
|
Map7
|
microtubule-associated protein 7 |
| chr14_-_114649173 | 0.11 |
ENSRNOT00000083528
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr14_-_82287706 | 0.10 |
ENSRNOT00000080695
|
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr7_-_83670356 | 0.10 |
ENSRNOT00000005584
|
Sybu
|
syntabulin |
| chr4_-_125929002 | 0.10 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr1_-_219640294 | 0.10 |
ENSRNOT00000054850
|
Kdm2a
|
lysine demethylase 2A |
| chr4_-_157358007 | 0.10 |
ENSRNOT00000090898
|
Gnb3
|
G protein subunit beta 3 |
| chr10_-_94488180 | 0.10 |
ENSRNOT00000015818
|
Gh1
|
growth hormone 1 |
| chr15_+_19547871 | 0.10 |
ENSRNOT00000036235
|
Gpr137c
|
G protein-coupled receptor 137C |
| chr13_-_73704480 | 0.09 |
ENSRNOT00000005296
|
Tor1aip1
|
torsin 1A interacting protein 1 |
| chr1_-_166912524 | 0.09 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr1_+_282188110 | 0.09 |
ENSRNOT00000013845
|
Fam45a
|
family with sequence similarity 45, member A |
| chr2_+_240668195 | 0.09 |
ENSRNOT00000087371
|
Manba
|
mannosidase beta |
| chr12_-_30151415 | 0.09 |
ENSRNOT00000079176
|
Crcp
|
CGRP receptor component |
| chr2_-_178297172 | 0.09 |
ENSRNOT00000038543
|
Fnip2
|
folliculin interacting protein 2 |
| chr15_+_39644851 | 0.09 |
ENSRNOT00000081373
|
Rcbtb1
|
RCC1 and BTB domain containing protein 1 |
| chr6_-_107080524 | 0.08 |
ENSRNOT00000011662
|
Zfyve1
|
zinc finger FYVE-type containing 1 |
| chr6_-_47890033 | 0.08 |
ENSRNOT00000011290
|
Colec11
|
collectin sub-family member 11 |
| chr7_-_123767797 | 0.08 |
ENSRNOT00000012699
|
Tcf20
|
transcription factor 20 |
| chr4_-_129544429 | 0.08 |
ENSRNOT00000091507
ENSRNOT00000083461 ENSRNOT00000087215 |
Tmf1
|
TATA element modulatory factor 1 |
| chr3_-_110324084 | 0.08 |
ENSRNOT00000010381
|
Bmf
|
Bcl2 modifying factor |
| chr9_+_81940630 | 0.08 |
ENSRNOT00000023067
|
Ttll4
|
tubulin tyrosine ligase like 4 |
| chr3_-_130114770 | 0.08 |
ENSRNOT00000010638
|
Jag1
|
jagged 1 |
| chrX_-_26376467 | 0.08 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr4_+_157126935 | 0.08 |
ENSRNOT00000056051
|
C1r
|
complement C1r |
| chr5_+_137371825 | 0.07 |
ENSRNOT00000072816
|
Tmem125
|
transmembrane protein 125 |
| chr2_+_77868412 | 0.07 |
ENSRNOT00000065897
ENSRNOT00000014022 |
Myo10
|
myosin X |
| chr3_+_177310753 | 0.07 |
ENSRNOT00000031448
|
Myt1
|
myelin transcription factor 1 |
| chr14_+_81488008 | 0.07 |
ENSRNOT00000018477
|
Tnip2
|
TNFAIP3 interacting protein 2 |
| chr1_-_214971929 | 0.07 |
ENSRNOT00000027195
|
Mob2
|
MOB kinase activator 2 |
| chr14_-_45797887 | 0.07 |
ENSRNOT00000029209
|
Tbc1d1
|
TBC1 domain family member 1 |
| chr3_-_33075685 | 0.07 |
ENSRNOT00000006937
|
Orc4
|
origin recognition complex, subunit 4 |
| chr5_-_154363329 | 0.07 |
ENSRNOT00000064596
|
Eloa
|
elongin A |
| chr18_-_27424090 | 0.06 |
ENSRNOT00000087968
|
Brd8
|
bromodomain containing 8 |
| chr2_+_197720259 | 0.06 |
ENSRNOT00000070919
|
Golph3l
|
golgi phosphoprotein 3-like |
| chr13_-_73704678 | 0.06 |
ENSRNOT00000005280
|
Tor1aip1
|
torsin 1A interacting protein 1 |
| chr8_-_96132634 | 0.06 |
ENSRNOT00000041655
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr6_-_105097054 | 0.06 |
ENSRNOT00000048606
|
Slc8a3
|
solute carrier family 8 member A3 |
| chr8_-_48582353 | 0.06 |
ENSRNOT00000011582
|
Pdzd3
|
PDZ domain containing 3 |
| chr9_-_120101098 | 0.06 |
ENSRNOT00000002917
|
Fam114a1l1
|
family with sequence similarity 114, member A1-like 1 |
| chr6_+_1410507 | 0.06 |
ENSRNOT00000034919
|
Gpatch11
|
G patch domain containing 11 |
| chr14_+_42007312 | 0.05 |
ENSRNOT00000063985
|
Atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr3_-_46153371 | 0.05 |
ENSRNOT00000085885
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr3_-_46051096 | 0.05 |
ENSRNOT00000081302
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr17_+_77167740 | 0.05 |
ENSRNOT00000042881
|
Optn
|
optineurin |
| chr2_-_227145185 | 0.05 |
ENSRNOT00000083189
|
AABR07013202.1
|
|
| chr12_+_41155497 | 0.05 |
ENSRNOT00000041741
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr7_-_139063752 | 0.05 |
ENSRNOT00000072309
|
LOC102551901
|
protein lifeguard 2-like |
| chr2_+_243840134 | 0.05 |
ENSRNOT00000093355
|
Eif4e
|
eukaryotic translation initiation factor 4E |
| chr16_+_6779281 | 0.05 |
ENSRNOT00000022865
|
Sfmbt1
|
Scm-like with four mbt domains 1 |
| chr7_-_2961873 | 0.05 |
ENSRNOT00000067441
|
Rps15-ps2
|
ribosomal protein S15, pseudogene 2 |
| chr3_-_64024205 | 0.05 |
ENSRNOT00000037015
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr1_+_217459215 | 0.05 |
ENSRNOT00000092386
ENSRNOT00000092357 |
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr9_-_32019205 | 0.05 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr7_+_64672722 | 0.05 |
ENSRNOT00000064448
ENSRNOT00000005539 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr2_-_53827175 | 0.05 |
ENSRNOT00000078158
|
RGD1305938
|
similar to expressed sequence AW549877 |
| chr4_+_147455533 | 0.05 |
ENSRNOT00000081957
|
Tsen2
|
tRNA splicing endonuclease subunit 2 |
| chr4_+_159483131 | 0.05 |
ENSRNOT00000089513
|
RGD1311164
|
similar to DNA segment, Chr 6, Wayne State University 163, expressed |
| chr10_-_47164596 | 0.05 |
ENSRNOT00000075751
|
LOC100911674
|
protein GTLF3B-like |
| chr4_-_50860756 | 0.04 |
ENSRNOT00000068404
|
Cadps2
|
calcium dependent secretion activator 2 |
| chr3_-_152159749 | 0.04 |
ENSRNOT00000067490
|
Cpne1
|
copine 1 |
| chr13_+_89597138 | 0.04 |
ENSRNOT00000004662
|
Apoa2
|
apolipoprotein A2 |
| chr18_-_5314511 | 0.04 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr12_+_40553741 | 0.04 |
ENSRNOT00000057396
|
LOC100360238
|
rCG21419-like |
| chr6_-_26624092 | 0.04 |
ENSRNOT00000008113
|
Trim54
|
tripartite motif-containing 54 |
| chr18_-_33434775 | 0.04 |
ENSRNOT00000040877
|
Yipf5
|
Yip1 domain family, member 5 |
| chr5_+_156587093 | 0.04 |
ENSRNOT00000019696
|
Hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr11_+_84745904 | 0.04 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr18_+_36596585 | 0.04 |
ENSRNOT00000036613
|
Rbm27
|
RNA binding motif protein 27 |
| chr17_+_1679627 | 0.04 |
ENSRNOT00000025801
|
Habp4
|
hyaluronan binding protein 4 |
| chr18_-_56870801 | 0.04 |
ENSRNOT00000087188
ENSRNOT00000032189 |
Arhgef37
|
Rho guanine nucleotide exchange factor 37 |
| chr5_-_33892462 | 0.04 |
ENSRNOT00000009334
|
Atp6v0d2
|
ATPase H+ transporting V0 subunit D2 |
| chr4_-_157155609 | 0.04 |
ENSRNOT00000016330
|
C1s
|
complement C1s |
| chr20_+_3351303 | 0.04 |
ENSRNOT00000080419
ENSRNOT00000001065 ENSRNOT00000086503 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr5_+_24860153 | 0.03 |
ENSRNOT00000042810
|
RGD1559441
|
similar to MIC2L1 |
| chr2_+_198683159 | 0.03 |
ENSRNOT00000028793
|
Txnip
|
thioredoxin interacting protein |
| chr6_+_58467254 | 0.03 |
ENSRNOT00000065396
|
Etv1
|
ets variant 1 |
| chr6_-_1410339 | 0.03 |
ENSRNOT00000035905
|
Heatr5b
|
HEAT repeat containing 5B |
| chr15_+_110114148 | 0.03 |
ENSRNOT00000006264
|
Itgbl1
|
integrin subunit beta like 1 |
| chr10_-_89338739 | 0.03 |
ENSRNOT00000073923
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr7_+_11838639 | 0.03 |
ENSRNOT00000025878
|
Ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr3_+_111097326 | 0.03 |
ENSRNOT00000018717
|
Vps18
|
VPS18 CORVET/HOPS core subunit |
| chr17_-_79085076 | 0.03 |
ENSRNOT00000057851
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr5_-_157759322 | 0.03 |
ENSRNOT00000023896
|
Pqlc2
|
PQ loop repeat containing 2 |
| chr14_-_37871051 | 0.03 |
ENSRNOT00000003087
|
Slain2
|
SLAIN motif family, member 2 |
| chr12_-_41993957 | 0.03 |
ENSRNOT00000001891
|
Rbm19
|
RNA binding motif protein 19 |
| chr1_+_171793782 | 0.03 |
ENSRNOT00000081351
|
Olfml1
|
olfactomedin-like 1 |
| chr6_+_137164535 | 0.02 |
ENSRNOT00000018225
|
Inf2
|
inverted formin, FH2 and WH2 domain containing |
| chr12_+_42343123 | 0.02 |
ENSRNOT00000043279
|
Oas1i
|
2 ' -5 ' oligoadenylate synthetase 1I |
| chr19_+_52032886 | 0.02 |
ENSRNOT00000019923
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr4_+_77519028 | 0.02 |
ENSRNOT00000008743
|
Zfp398
|
zinc finger protein 398 |
| chr14_-_17436897 | 0.02 |
ENSRNOT00000003277
|
Uso1
|
USO1 vesicle transport factor |
| chr5_+_154522119 | 0.02 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr3_-_171286413 | 0.02 |
ENSRNOT00000008365
ENSRNOT00000081036 |
Zbp1
|
Z-DNA binding protein 1 |
| chr15_-_4022223 | 0.02 |
ENSRNOT00000081997
|
Zswim8
|
zinc finger, SWIM-type containing 8 |
| chrX_+_71342775 | 0.02 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
| chr5_+_140712583 | 0.02 |
ENSRNOT00000019587
|
Trit1
|
tRNA isopentenyltransferase 1 |
| chr4_+_114876770 | 0.02 |
ENSRNOT00000068102
ENSRNOT00000078175 |
Dctn1
|
dynactin subunit 1 |
| chr10_+_105796680 | 0.02 |
ENSRNOT00000000264
|
Mfsd11
|
major facilitator superfamily domain containing 11 |
| chr10_+_11206226 | 0.02 |
ENSRNOT00000006979
|
Tfap4
|
transcription factor AP-4 |
| chr9_-_92435363 | 0.01 |
ENSRNOT00000093735
ENSRNOT00000022822 ENSRNOT00000093245 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chr14_+_91588940 | 0.01 |
ENSRNOT00000057067
|
RGD1309870
|
hypothetical LOC289778 |
| chr10_+_99437436 | 0.01 |
ENSRNOT00000006254
|
Kcnj2
|
potassium voltage-gated channel subfamily J member 2 |
| chrX_-_119162518 | 0.01 |
ENSRNOT00000073176
|
LOC103694506
|
ubiquitin-conjugating enzyme E2 W |
| chr17_-_8429338 | 0.01 |
ENSRNOT00000016390
|
Tgfbi
|
transforming growth factor, beta induced |
| chr2_-_2779109 | 0.01 |
ENSRNOT00000017098
|
Rfesd
|
Rieske (Fe-S) domain containing |
| chr17_-_29360822 | 0.01 |
ENSRNOT00000080031
|
Fars2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr11_+_47061354 | 0.01 |
ENSRNOT00000039997
|
Pcnp
|
PEST proteolytic signal containing nuclear protein |
| chr17_-_45286923 | 0.01 |
ENSRNOT00000041103
|
AABR07027819.1
|
|
| chrX_-_119532965 | 0.01 |
ENSRNOT00000073677
|
AABR07041109.1
|
|
| chr5_+_172364421 | 0.01 |
ENSRNOT00000018769
|
Hes5
|
hes family bHLH transcription factor 5 |
| chr9_+_17705212 | 0.01 |
ENSRNOT00000026728
|
Tmem63b
|
transmembrane protein 63B |
| chr7_+_18440742 | 0.01 |
ENSRNOT00000011513
|
Myo1f
|
myosin IF |
| chr19_+_24329023 | 0.01 |
ENSRNOT00000065558
|
Tbc1d9
|
TBC1 domain family member 9 |
| chr5_+_151692108 | 0.01 |
ENSRNOT00000086144
|
Fam46b
|
family with sequence similarity 46, member B |
| chr1_-_273758442 | 0.01 |
ENSRNOT00000088016
|
Xpnpep1
|
X-prolyl aminopeptidase 1 |
| chr9_+_81821346 | 0.01 |
ENSRNOT00000022234
|
Plcd4
|
phospholipase C, delta 4 |
| chr2_-_210943620 | 0.01 |
ENSRNOT00000026750
|
Gpr61
|
G protein-coupled receptor 61 |
| chr2_-_184289126 | 0.01 |
ENSRNOT00000081678
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr4_+_62380914 | 0.01 |
ENSRNOT00000029845
|
Tmem140
|
transmembrane protein 140 |
| chr7_-_60743328 | 0.01 |
ENSRNOT00000066767
|
Mdm2
|
MDM2 proto-oncogene |
| chr3_+_7422820 | 0.01 |
ENSRNOT00000064323
|
Ddx31
|
DEAD-box helicase 31 |
| chr1_-_89509343 | 0.00 |
ENSRNOT00000028637
|
Fxyd3
|
FXYD domain-containing ion transport regulator 3 |
| chr10_-_56289882 | 0.00 |
ENSRNOT00000090762
ENSRNOT00000056903 |
Tnfsf13
|
tumor necrosis factor superfamily member 13 |
| chr2_-_94730978 | 0.00 |
ENSRNOT00000087598
|
Zbtb10
|
zinc finger and BTB domain containing 10 |
| chr19_+_14392423 | 0.00 |
ENSRNOT00000018880
|
Tom1
|
target of myb1 membrane trafficking protein |
| chr1_-_273758247 | 0.00 |
ENSRNOT00000033148
|
Xpnpep1
|
X-prolyl aminopeptidase 1 |
| chr2_+_32820322 | 0.00 |
ENSRNOT00000013768
|
Cd180
|
CD180 molecule |
| chr20_-_20680675 | 0.00 |
ENSRNOT00000087085
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr1_+_100224401 | 0.00 |
ENSRNOT00000025846
|
Fv1
|
Friend virus susceptibility 1 |
| chr1_+_201055644 | 0.00 |
ENSRNOT00000054937
ENSRNOT00000047161 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr4_+_24612205 | 0.00 |
ENSRNOT00000057382
|
Ewsr1
|
EWS RNA-binding protein 1 |
| chr2_+_189423559 | 0.00 |
ENSRNOT00000029076
|
Tpm3
|
tropomyosin 3 |
| chr8_-_39734594 | 0.00 |
ENSRNOT00000076872
|
Slc37a2
|
solute carrier family 37 member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.3 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.4 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.1 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.1 | GO:0061443 | endocardial cushion cell differentiation(GO:0061443) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:1904116 | response to vasopressin(GO:1904116) |
| 0.0 | 0.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.1 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.0 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.0 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.0 | 0.2 | GO:2000641 | regulation of early endosome to late endosome transport(GO:2000641) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.2 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0032437 | spectrin(GO:0008091) cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0016842 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0001635 | calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |