Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
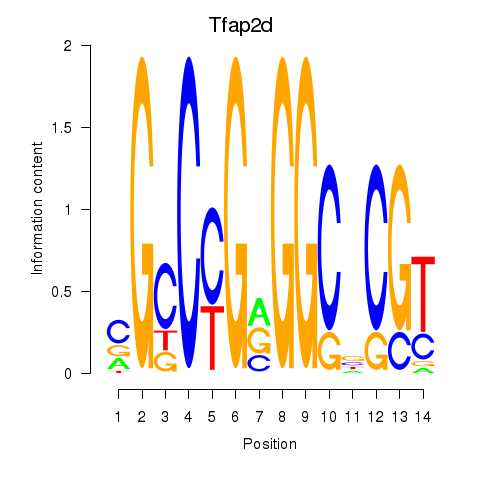
Results for Tfap2d
Z-value: 0.39
Transcription factors associated with Tfap2d
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2d
|
ENSRNOG00000011642 | transcription factor AP-2 delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2d | rn6_v1_chr9_+_25304876_25304876 | 0.19 | 7.6e-01 | Click! |
Activity profile of Tfap2d motif
Sorted Z-values of Tfap2d motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_226501920 | 0.13 |
ENSRNOT00000050716
|
Lrrc10b
|
leucine rich repeat containing 10B |
| chr8_+_53678777 | 0.12 |
ENSRNOT00000045944
|
Drd2
|
dopamine receptor D2 |
| chr1_-_79930263 | 0.08 |
ENSRNOT00000019219
|
Foxa3
|
forkhead box A3 |
| chr10_-_56403188 | 0.08 |
ENSRNOT00000019947
|
Chrnb1
|
cholinergic receptor nicotinic beta 1 subunit |
| chr10_-_85049331 | 0.08 |
ENSRNOT00000012538
|
Tbx21
|
T-box 21 |
| chr9_+_99998275 | 0.06 |
ENSRNOT00000074395
|
Gpc1
|
glypican 1 |
| chr4_-_113610243 | 0.06 |
ENSRNOT00000008813
|
Hk2
|
hexokinase 2 |
| chr14_-_81725181 | 0.06 |
ENSRNOT00000078252
|
Cfap99
|
cilia and flagella associated protein 99 |
| chr13_+_51795867 | 0.05 |
ENSRNOT00000006747
|
Ube2t
|
ubiquitin-conjugating enzyme E2T |
| chr3_-_171342646 | 0.05 |
ENSRNOT00000071853
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr6_+_107485249 | 0.05 |
ENSRNOT00000080859
|
Acot1
|
acyl-CoA thioesterase 1 |
| chr3_+_13838304 | 0.05 |
ENSRNOT00000025067
|
Hspa5
|
heat shock protein family A member 5 |
| chr3_-_15433252 | 0.05 |
ENSRNOT00000008817
|
Lhx6
|
LIM homeobox 6 |
| chr11_+_87549059 | 0.04 |
ENSRNOT00000002567
|
Ccdc74a
|
coiled-coil domain containing 74A |
| chr2_+_165486910 | 0.04 |
ENSRNOT00000012982
|
LOC100362176
|
hypothetical protein LOC100362176 |
| chr6_+_137243185 | 0.04 |
ENSRNOT00000030879
|
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr5_-_61077627 | 0.04 |
ENSRNOT00000015338
|
Shb
|
SH2 domain containing adaptor protein B |
| chr7_+_116745061 | 0.04 |
ENSRNOT00000076174
|
Rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr1_-_270472866 | 0.04 |
ENSRNOT00000015979
|
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr1_+_198690794 | 0.03 |
ENSRNOT00000023999
|
Zfp771
|
zinc finger protein 771 |
| chr10_+_89181180 | 0.03 |
ENSRNOT00000078931
ENSRNOT00000027770 |
Wnk4
|
WNK lysine deficient protein kinase 4 |
| chr2_-_186245342 | 0.03 |
ENSRNOT00000057062
ENSRNOT00000022292 |
Dclk2
|
doublecortin-like kinase 2 |
| chr1_-_254671778 | 0.03 |
ENSRNOT00000025493
|
Htr7
|
5-hydroxytryptamine receptor 7 |
| chr1_-_141349881 | 0.03 |
ENSRNOT00000021109
|
Rhcg
|
Rh family, C glycoprotein |
| chr4_-_153028129 | 0.03 |
ENSRNOT00000074558
|
Cecr6
|
cat eye syndrome chromosome region, candidate 6 |
| chr14_+_11198896 | 0.03 |
ENSRNOT00000079767
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr1_-_92069160 | 0.02 |
ENSRNOT00000039115
|
Dpy19l3
|
dpy-19-like 3 (C. elegans) |
| chr8_-_63092009 | 0.02 |
ENSRNOT00000034300
|
Loxl1
|
lysyl oxidase-like 1 |
| chr5_+_148193710 | 0.02 |
ENSRNOT00000088568
|
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr3_-_79390956 | 0.02 |
ENSRNOT00000077943
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr18_+_25163561 | 0.02 |
ENSRNOT00000042287
ENSRNOT00000049898 ENSRNOT00000017573 |
Bin1
|
bridging integrator 1 |
| chr6_+_28571351 | 0.02 |
ENSRNOT00000005389
|
Adcy3
|
adenylate cyclase 3 |
| chr3_+_111545007 | 0.02 |
ENSRNOT00000007247
|
Itpka
|
inositol-trisphosphate 3-kinase A |
| chr1_+_46378807 | 0.02 |
ENSRNOT00000043521
|
Zdhhc14
|
zinc finger, DHHC-type containing 14 |
| chr12_-_17358617 | 0.02 |
ENSRNOT00000001733
|
Gpr146
|
G protein-coupled receptor 146 |
| chr14_-_11198194 | 0.02 |
ENSRNOT00000003083
|
Enoph1
|
enolase-phosphatase 1 |
| chr4_+_145580799 | 0.02 |
ENSRNOT00000013727
|
Vhl
|
von Hippel-Lindau tumor suppressor |
| chr10_-_11174861 | 0.02 |
ENSRNOT00000006680
|
Glis2
|
GLIS family zinc finger 2 |
| chr2_-_183031214 | 0.02 |
ENSRNOT00000013260
|
LOC679811
|
similar to RIKEN cDNA D930015E06 |
| chr3_+_114747410 | 0.02 |
ENSRNOT00000043414
|
Spata5l1
|
spermatogenesis associated 5-like 1 |
| chr8_+_116311078 | 0.02 |
ENSRNOT00000037363
|
Rassf1
|
Ras association domain family member 1 |
| chr1_+_154377247 | 0.02 |
ENSRNOT00000092945
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chrX_-_114232939 | 0.02 |
ENSRNOT00000042639
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr3_+_164822111 | 0.02 |
ENSRNOT00000014568
|
Pard6b
|
par-6 family cell polarity regulator beta |
| chr3_+_159421671 | 0.02 |
ENSRNOT00000010343
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr1_+_84470829 | 0.02 |
ENSRNOT00000025472
|
Ttc9b
|
tetratricopeptide repeat domain 9B |
| chr1_-_146289465 | 0.02 |
ENSRNOT00000017362
|
Abhd17c
|
abhydrolase domain containing 17C |
| chr1_+_72580424 | 0.02 |
ENSRNOT00000022977
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chrX_+_127562660 | 0.02 |
ENSRNOT00000029031
ENSRNOT00000010367 |
Gria3
|
glutamate ionotropic receptor AMPA type subunit 3 |
| chr1_-_179010257 | 0.02 |
ENSRNOT00000017199
|
Rras2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr1_-_254671596 | 0.02 |
ENSRNOT00000025450
|
Htr7
|
5-hydroxytryptamine receptor 7 |
| chr2_-_31826867 | 0.02 |
ENSRNOT00000025687
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr8_+_128087345 | 0.02 |
ENSRNOT00000019777
|
Acvr2b
|
activin A receptor type 2B |
| chr4_-_145147397 | 0.02 |
ENSRNOT00000010347
|
Lhfpl4
|
lipoma HMGIC fusion partner-like 4 |
| chr1_+_261389804 | 0.02 |
ENSRNOT00000064160
|
Marveld1
|
MARVEL domain containing 1 |
| chr12_+_31934343 | 0.02 |
ENSRNOT00000011181
|
Tmem132d
|
transmembrane protein 132D |
| chr1_-_80515694 | 0.01 |
ENSRNOT00000075144
|
Clasrp
|
CLK4-associating serine/arginine rich protein |
| chr1_+_82480195 | 0.01 |
ENSRNOT00000028051
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr8_+_115069095 | 0.01 |
ENSRNOT00000014770
|
Dusp7
|
dual specificity phosphatase 7 |
| chr19_+_24747178 | 0.01 |
ENSRNOT00000038879
ENSRNOT00000079106 |
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr16_+_62483761 | 0.01 |
ENSRNOT00000058805
|
Wrn
|
Werner syndrome RecQ like helicase |
| chr8_-_116391158 | 0.01 |
ENSRNOT00000078720
ENSRNOT00000022550 |
Gnai2
|
G protein subunit alpha i2 |
| chr3_+_2591331 | 0.01 |
ENSRNOT00000017466
|
Sapcd2
|
suppressor APC domain containing 2 |
| chr14_+_39661968 | 0.01 |
ENSRNOT00000064779
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chrX_+_156963870 | 0.01 |
ENSRNOT00000077109
|
Pdzd4
|
PDZ domain containing 4 |
| chr5_-_156689415 | 0.01 |
ENSRNOT00000083474
|
Pink1
|
PTEN induced putative kinase 1 |
| chr12_-_17972737 | 0.01 |
ENSRNOT00000001783
|
Fam20c
|
FAM20C, golgi associated secretory pathway kinase |
| chr11_-_61530567 | 0.01 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr19_+_21372163 | 0.01 |
ENSRNOT00000020329
|
Siah1
|
siah E3 ubiquitin protein ligase 1 |
| chr1_+_267359830 | 0.01 |
ENSRNOT00000015495
|
Slk
|
STE20-like kinase |
| chr4_-_58250798 | 0.01 |
ENSRNOT00000048436
|
Klf14
|
Kruppel-like factor 14 |
| chr2_-_186245163 | 0.01 |
ENSRNOT00000089339
|
Dclk2
|
doublecortin-like kinase 2 |
| chr10_-_85222861 | 0.01 |
ENSRNOT00000031045
|
Npepps
|
aminopeptidase puromycin sensitive |
| chr19_+_52032886 | 0.01 |
ENSRNOT00000019923
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr8_+_61659445 | 0.01 |
ENSRNOT00000023831
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chrX_-_32355296 | 0.01 |
ENSRNOT00000081652
ENSRNOT00000065075 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr4_+_157511642 | 0.01 |
ENSRNOT00000065846
|
Pianp
|
PILR alpha associated neural protein |
| chr2_+_151685251 | 0.01 |
ENSRNOT00000019340
|
Rap2b
|
RAP2B, member of RAS oncogene family |
| chr18_-_18079560 | 0.01 |
ENSRNOT00000072093
|
AABR07031533.1
|
|
| chr1_-_125967756 | 0.01 |
ENSRNOT00000031488
|
Fam189a1
|
family with sequence similarity 189, member A1 |
| chr1_+_80056755 | 0.01 |
ENSRNOT00000021221
|
Snrpd2
|
small nuclear ribonucleoprotein D2 polypeptide |
| chr17_+_11953552 | 0.01 |
ENSRNOT00000090782
|
Ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr11_+_64601029 | 0.01 |
ENSRNOT00000004138
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chrX_-_33665821 | 0.01 |
ENSRNOT00000066676
|
Rbbp7
|
RB binding protein 7, chromatin remodeling factor |
| chr6_+_93740586 | 0.01 |
ENSRNOT00000011466
|
Dact1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr11_-_34598275 | 0.01 |
ENSRNOT00000077233
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr8_+_115179893 | 0.01 |
ENSRNOT00000017469
|
Rrp9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr19_-_11669578 | 0.01 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr12_+_15890170 | 0.01 |
ENSRNOT00000001654
|
Gna12
|
G protein subunit alpha 12 |
| chr7_+_130474508 | 0.01 |
ENSRNOT00000085191
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr3_-_11747113 | 0.01 |
ENSRNOT00000032893
|
Cdk9
|
cyclin-dependent kinase 9 |
| chrX_+_63343546 | 0.01 |
ENSRNOT00000076315
|
Klhl15
|
kelch-like family member 15 |
| chr19_+_37830917 | 0.01 |
ENSRNOT00000025608
|
Nutf2
|
nuclear transport factor 2 |
| chr3_+_152933771 | 0.01 |
ENSRNOT00000027539
|
RGD1307752
|
similar to RIKEN cDNA 1110008F13 |
| chr6_-_96442682 | 0.01 |
ENSRNOT00000010923
|
Tmem30b
|
transmembrane protein 30B |
| chr5_+_147714163 | 0.01 |
ENSRNOT00000012663
|
Marcksl1
|
MARCKS-like 1 |
| chr19_-_55300403 | 0.01 |
ENSRNOT00000018591
|
Rnf166
|
ring finger protein 166 |
| chr19_+_34139997 | 0.01 |
ENSRNOT00000017867
|
Arhgap10
|
Rho GTPase activating protein 10 |
| chr10_+_45659143 | 0.01 |
ENSRNOT00000058327
|
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr19_+_53635688 | 0.01 |
ENSRNOT00000067141
|
Map1lc3b
|
microtubule-associated protein 1 light chain 3 beta |
| chr7_-_70498992 | 0.01 |
ENSRNOT00000067774
ENSRNOT00000079327 |
Pip4k2c
|
phosphatidylinositol-5-phosphate 4-kinase type 2 gamma |
| chr6_-_137733026 | 0.01 |
ENSRNOT00000019213
|
Jag2
|
jagged 2 |
| chr8_+_116154913 | 0.01 |
ENSRNOT00000081242
|
Cacna2d2
|
calcium voltage-gated channel auxiliary subunit alpha2delta 2 |
| chr3_+_79678201 | 0.01 |
ENSRNOT00000087604
ENSRNOT00000079709 |
Mtch2
|
mitochondrial carrier 2 |
| chr13_+_97702097 | 0.01 |
ENSRNOT00000057787
|
LOC103689999
|
saccharopine dehydrogenase-like oxidoreductase |
| chr9_-_82214440 | 0.01 |
ENSRNOT00000024419
|
Ihh
|
Indian hedgehog |
| chr16_+_83824430 | 0.01 |
ENSRNOT00000032918
|
Irs2
|
insulin receptor substrate 2 |
| chr1_+_101554642 | 0.01 |
ENSRNOT00000028474
|
Bcat2
|
branched chain amino acid transaminase 2 |
| chr20_-_6960481 | 0.01 |
ENSRNOT00000093172
|
Mtch1
|
mitochondrial carrier 1 |
| chr8_+_116154736 | 0.01 |
ENSRNOT00000021218
|
Cacna2d2
|
calcium voltage-gated channel auxiliary subunit alpha2delta 2 |
| chr5_+_8666200 | 0.01 |
ENSRNOT00000007766
|
Arfgef1
|
ADP ribosylation factor guanine nucleotide exchange factor 1 |
| chr6_+_137824213 | 0.01 |
ENSRNOT00000056880
|
Pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr4_+_70903253 | 0.01 |
ENSRNOT00000019720
|
Ephb6
|
Eph receptor B6 |
| chr11_-_34598102 | 0.01 |
ENSRNOT00000068743
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr19_-_25801526 | 0.01 |
ENSRNOT00000003884
|
Nacc1
|
nucleus accumbens associated 1 |
| chr16_-_21274688 | 0.01 |
ENSRNOT00000027953
|
Tssk6
|
testis-specific serine kinase 6 |
| chr10_+_86711240 | 0.01 |
ENSRNOT00000012812
|
Msl1
|
male specific lethal 1 homolog |
| chr10_+_85744568 | 0.01 |
ENSRNOT00000005522
|
Lasp1
|
LIM and SH3 protein 1 |
| chr16_+_4469468 | 0.01 |
ENSRNOT00000021164
|
Wnt5a
|
wingless-type MMTV integration site family, member 5A |
| chr6_+_107596782 | 0.00 |
ENSRNOT00000076882
|
Dnal1
|
dynein, axonemal, light chain 1 |
| chrX_+_54734385 | 0.00 |
ENSRNOT00000005023
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr10_-_59360661 | 0.00 |
ENSRNOT00000036942
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr5_-_151117042 | 0.00 |
ENSRNOT00000066549
|
Fam76a
|
family with sequence similarity 76, member A |
| chr2_+_3400977 | 0.00 |
ENSRNOT00000093593
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr16_+_61954590 | 0.00 |
ENSRNOT00000017883
|
Rbpms
|
RNA binding protein with multiple splicing |
| chr11_-_44292884 | 0.00 |
ENSRNOT00000084060
|
Dcbld2
|
discoidin, CUB and LCCL domain containing 2 |
| chr4_-_66624912 | 0.00 |
ENSRNOT00000064891
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr2_+_24449485 | 0.00 |
ENSRNOT00000073688
|
Tbca
|
tubulin folding cofactor A |
| chr1_+_143036218 | 0.00 |
ENSRNOT00000025797
|
Pde8a
|
phosphodiesterase 8A |
| chr3_+_147772165 | 0.00 |
ENSRNOT00000008713
|
Tbc1d20
|
TBC1 domain family, member 20 |
| chr3_-_148706972 | 0.00 |
ENSRNOT00000036956
|
Tspy26
|
testis specific protein, Y-linked 26 |
| chr16_-_71237118 | 0.00 |
ENSRNOT00000066974
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr3_+_175408629 | 0.00 |
ENSRNOT00000081344
|
Lsm14b
|
LSM family member 14B |
| chr1_+_200037749 | 0.00 |
ENSRNOT00000027630
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr3_+_6211789 | 0.00 |
ENSRNOT00000012892
|
Rxra
|
retinoid X receptor alpha |
| chr9_+_10846596 | 0.00 |
ENSRNOT00000075386
|
Dpp9
|
dipeptidyl peptidase 9 |
| chr1_-_220480132 | 0.00 |
ENSRNOT00000027421
|
Cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr20_-_4698718 | 0.00 |
ENSRNOT00000047527
|
RT1-CE7
|
RT1 class I, locus CE7 |
| chr9_+_66716969 | 0.00 |
ENSRNOT00000031261
|
Fam117b
|
family with sequence similarity 117, member B |
| chr1_+_214317682 | 0.00 |
ENSRNOT00000032904
|
Tmem80
|
transmembrane protein 80 |
| chr2_-_43999107 | 0.00 |
ENSRNOT00000065473
|
LOC100912399
|
mitogen-activated protein kinase kinase kinase 1-like |
| chr8_+_50310405 | 0.00 |
ENSRNOT00000073507
|
Sik3
|
SIK family kinase 3 |
| chr17_+_36334589 | 0.00 |
ENSRNOT00000081368
|
E2f3
|
E2F transcription factor 3 |
| chr3_+_11317183 | 0.00 |
ENSRNOT00000091171
ENSRNOT00000016341 |
Golga2
|
golgin A2 |
| chr7_-_139394166 | 0.00 |
ENSRNOT00000082429
|
Vdr
|
vitamin D (1,25- dihydroxyvitamin D3) receptor |
| chr1_+_79911516 | 0.00 |
ENSRNOT00000019075
|
Irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr2_+_95320283 | 0.00 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2d
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051944 | glial cell-derived neurotrophic factor secretion(GO:0044467) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) negative regulation of dopamine receptor signaling pathway(GO:0060160) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.0 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 | 0.1 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.1 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.0 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |