Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
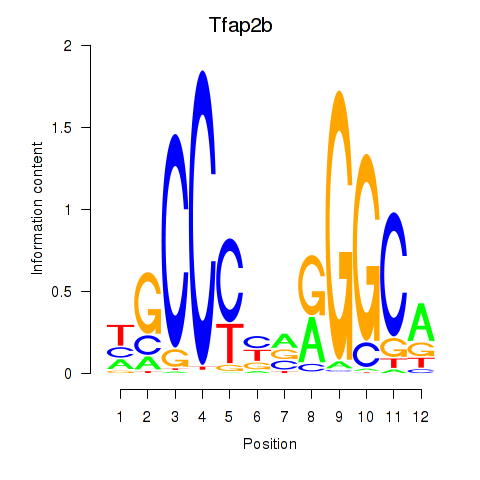
Results for Tfap2b
Z-value: 0.31
Transcription factors associated with Tfap2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2b
|
ENSRNOG00000011823 | transcription factor AP-2 beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2b | rn6_v1_chr9_+_25410669_25410717 | -0.08 | 8.9e-01 | Click! |
Activity profile of Tfap2b motif
Sorted Z-values of Tfap2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_123179644 | 0.15 |
ENSRNOT00000028835
|
Lzts3
|
leucine zipper tumor suppressor family member 3 |
| chr3_+_155160481 | 0.14 |
ENSRNOT00000021133
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr6_-_99783047 | 0.12 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr10_-_85684138 | 0.11 |
ENSRNOT00000017989
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase type 2 beta |
| chr17_+_70684539 | 0.11 |
ENSRNOT00000025700
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr17_+_70684134 | 0.11 |
ENSRNOT00000025731
ENSRNOT00000068354 |
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr1_+_78876205 | 0.10 |
ENSRNOT00000022610
|
Pnmal2
|
paraneoplastic Ma antigen family-like 2 |
| chr19_+_46761570 | 0.09 |
ENSRNOT00000058779
|
Wwox
|
WW domain-containing oxidoreductase |
| chr1_+_87045796 | 0.09 |
ENSRNOT00000027620
|
Lgals7
|
galectin 7 |
| chr7_+_130498199 | 0.09 |
ENSRNOT00000092684
ENSRNOT00000092431 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr1_+_261337594 | 0.09 |
ENSRNOT00000019874
|
Pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr5_+_63781801 | 0.09 |
ENSRNOT00000008302
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr1_-_219422268 | 0.09 |
ENSRNOT00000025092
|
Carns1
|
carnosine synthase 1 |
| chr2_+_189629297 | 0.08 |
ENSRNOT00000049810
|
Dennd4b
|
DENN domain containing 4B |
| chr10_+_30833914 | 0.07 |
ENSRNOT00000035346
ENSRNOT00000079437 |
Clint1
|
clathrin interactor 1 |
| chr1_-_142164007 | 0.07 |
ENSRNOT00000078982
|
Man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chr7_-_140546908 | 0.07 |
ENSRNOT00000077502
|
Kmt2d
|
lysine methyltransferase 2D |
| chr19_-_55490426 | 0.07 |
ENSRNOT00000081800
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr4_+_123801174 | 0.07 |
ENSRNOT00000029055
|
RGD1560289
|
similar to chromosome 3 open reading frame 20 |
| chr11_-_83926524 | 0.06 |
ENSRNOT00000041777
ENSRNOT00000040029 |
Eif4g1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr8_+_48805684 | 0.06 |
ENSRNOT00000064041
|
Bcl9l
|
B-cell CLL/lymphoma 9-like |
| chr5_+_150833819 | 0.06 |
ENSRNOT00000056170
|
Eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr7_-_118332577 | 0.06 |
ENSRNOT00000090370
|
Rbfox2
|
RNA binding protein, fox-1 homolog 2 |
| chr8_-_131899023 | 0.06 |
ENSRNOT00000034690
|
Zfp445
|
zinc finger protein 445 |
| chr20_+_3823596 | 0.06 |
ENSRNOT00000087670
|
Rxrb
|
retinoid X receptor beta |
| chr5_+_59348639 | 0.06 |
ENSRNOT00000084031
ENSRNOT00000060264 |
Reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr5_-_155381732 | 0.06 |
ENSRNOT00000033670
|
Zbtb40
|
zinc finger and BTB domain containing 40 |
| chr4_-_150616895 | 0.05 |
ENSRNOT00000073562
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr20_+_3827367 | 0.05 |
ENSRNOT00000079967
|
Rxrb
|
retinoid X receptor beta |
| chr4_-_88649216 | 0.05 |
ENSRNOT00000058626
|
Herc6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr17_-_10208360 | 0.05 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr1_-_142164263 | 0.05 |
ENSRNOT00000016281
|
Man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chrX_+_68782872 | 0.05 |
ENSRNOT00000075995
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr6_+_26566494 | 0.05 |
ENSRNOT00000079292
|
Gtf3c2
|
general transcription factor IIIC subunit 2 |
| chr3_-_15433252 | 0.05 |
ENSRNOT00000008817
|
Lhx6
|
LIM homeobox 6 |
| chr2_+_116028781 | 0.05 |
ENSRNOT00000089477
|
Phc3
|
polyhomeotic homolog 3 |
| chr18_-_68551558 | 0.05 |
ENSRNOT00000016369
|
Rab27b
|
RAB27B, member RAS oncogene family |
| chr1_+_222519615 | 0.05 |
ENSRNOT00000083585
|
Rcor2
|
REST corepressor 2 |
| chr19_+_37568113 | 0.05 |
ENSRNOT00000023710
|
Fam65a
|
family with sequence similarity 65, member A |
| chr7_-_116781766 | 0.04 |
ENSRNOT00000010084
|
Mafa
|
MAF bZIP transcription factor A |
| chr9_+_54212767 | 0.04 |
ENSRNOT00000078073
ENSRNOT00000090026 |
Gls
|
glutaminase |
| chr1_+_154377447 | 0.04 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr10_+_17421075 | 0.04 |
ENSRNOT00000047011
|
Stk10
|
serine/threonine kinase 10 |
| chr20_-_31072469 | 0.04 |
ENSRNOT00000082448
|
Eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr5_-_136965191 | 0.04 |
ENSRNOT00000056842
|
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr10_-_73787083 | 0.04 |
ENSRNOT00000035280
|
Med13
|
mediator complex subunit 13 |
| chr3_+_3310954 | 0.04 |
ENSRNOT00000061773
|
Kcnt1
|
potassium sodium-activated channel subfamily T member 1 |
| chr14_+_64686793 | 0.04 |
ENSRNOT00000005894
ENSRNOT00000036646 |
Adgra3
|
adhesion G protein-coupled receptor A3 |
| chr1_-_48891130 | 0.04 |
ENSRNOT00000083884
|
AC135026.1
|
|
| chr9_+_16749809 | 0.04 |
ENSRNOT00000028786
|
Cul9
|
cullin 9 |
| chr17_+_76306585 | 0.04 |
ENSRNOT00000065978
|
Dhtkd1
|
dehydrogenase E1 and transketolase domain containing 1 |
| chr10_+_13145099 | 0.04 |
ENSRNOT00000091287
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr10_-_62287189 | 0.04 |
ENSRNOT00000004365
|
Wdr81
|
WD repeat domain 81 |
| chr5_+_151436464 | 0.04 |
ENSRNOT00000012463
|
Map3k6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr19_-_55423052 | 0.04 |
ENSRNOT00000019528
ENSRNOT00000079641 |
Galns
|
galactosamine (N-acetyl)-6-sulfatase |
| chr12_-_30810964 | 0.03 |
ENSRNOT00000001235
ENSRNOT00000093237 ENSRNOT00000077977 |
Sfswap
|
splicing factor SWAP homolog |
| chr4_+_157523770 | 0.03 |
ENSRNOT00000055985
ENSRNOT00000023240 |
Zfp384
|
zinc finger protein 384 |
| chr15_-_28611946 | 0.03 |
ENSRNOT00000016288
|
Supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr8_-_111984572 | 0.03 |
ENSRNOT00000014519
|
Tmem108
|
transmembrane protein 108 |
| chr1_+_87045335 | 0.03 |
ENSRNOT00000084393
|
Lgals7
|
galectin 7 |
| chrX_+_120859968 | 0.03 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr20_+_5067330 | 0.03 |
ENSRNOT00000037482
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr15_+_19690194 | 0.03 |
ENSRNOT00000010612
|
Styx
|
serine/threonine/tyrosine interacting protein |
| chr12_-_37682964 | 0.03 |
ENSRNOT00000001421
|
RGD1563482
|
similar to hypothetical protein FLJ38663 |
| chr1_+_177764445 | 0.03 |
ENSRNOT00000037185
|
Rassf10
|
Ras association domain family member 10 |
| chr8_-_56696523 | 0.03 |
ENSRNOT00000080043
|
Zc3h12c
|
zinc finger CCCH type containing 12C |
| chr5_-_74191167 | 0.03 |
ENSRNOT00000088169
|
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr4_+_7662019 | 0.03 |
ENSRNOT00000014023
|
Fam126a
|
family with sequence similarity 126, member A |
| chrX_-_114232939 | 0.03 |
ENSRNOT00000042639
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr4_+_140092352 | 0.03 |
ENSRNOT00000008892
|
Setmar
|
SET domain and mariner transposase fusion gene |
| chr11_-_47113993 | 0.03 |
ENSRNOT00000034940
|
Zbtb11
|
zinc finger and BTB domain containing 11 |
| chr9_+_17086475 | 0.03 |
ENSRNOT00000025660
|
Tjap1
|
tight junction associated protein 1 |
| chr17_-_63994169 | 0.03 |
ENSRNOT00000075651
|
Chrm3
|
cholinergic receptor, muscarinic 3 |
| chr7_-_63578490 | 0.03 |
ENSRNOT00000007295
|
Rassf3
|
Ras association domain family member 3 |
| chr8_+_118821729 | 0.03 |
ENSRNOT00000028409
|
Setd2
|
SET domain containing 2 |
| chr1_-_162300515 | 0.03 |
ENSRNOT00000031161
|
Usp35
|
ubiquitin specific peptidase 35 |
| chr3_-_162581610 | 0.03 |
ENSRNOT00000079324
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr13_+_51795867 | 0.02 |
ENSRNOT00000006747
|
Ube2t
|
ubiquitin-conjugating enzyme E2T |
| chr8_-_56696968 | 0.02 |
ENSRNOT00000016637
|
Zc3h12c
|
zinc finger CCCH type containing 12C |
| chr3_-_57913999 | 0.02 |
ENSRNOT00000090426
|
Slc25a12
|
solute carrier family 25 member 12 |
| chr2_-_234296145 | 0.02 |
ENSRNOT00000014155
|
Elovl6
|
ELOVL fatty acid elongase 6 |
| chr1_+_1784078 | 0.02 |
ENSRNOT00000020098
|
Lats1
|
large tumor suppressor kinase 1 |
| chr17_-_9695292 | 0.02 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr1_-_127599257 | 0.02 |
ENSRNOT00000018436
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
| chrX_+_14498283 | 0.02 |
ENSRNOT00000092143
|
Xk
|
X-linked Kx blood group |
| chr13_-_72869396 | 0.02 |
ENSRNOT00000093563
|
RGD1304622
|
similar to 6820428L09 protein |
| chr3_+_79729739 | 0.02 |
ENSRNOT00000084833
|
Kbtbd4
|
kelch repeat and BTB domain containing 4 |
| chr7_-_36000906 | 0.02 |
ENSRNOT00000011178
|
Plxnc1
|
plexin C1 |
| chr8_+_63036976 | 0.02 |
ENSRNOT00000011383
|
Stoml1
|
stomatin like 1 |
| chr7_-_119185238 | 0.02 |
ENSRNOT00000007598
|
Foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr20_+_3556560 | 0.02 |
ENSRNOT00000085635
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr13_-_49169918 | 0.01 |
ENSRNOT00000000036
|
Tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr6_-_110904288 | 0.01 |
ENSRNOT00000014645
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr7_+_78092037 | 0.01 |
ENSRNOT00000050753
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr13_-_53108713 | 0.01 |
ENSRNOT00000035404
|
RGD1311892
|
similar to hypothetical protein FLJ10901 |
| chr16_-_61091169 | 0.01 |
ENSRNOT00000016328
|
Dusp4
|
dual specificity phosphatase 4 |
| chr1_+_221735517 | 0.01 |
ENSRNOT00000028628
ENSRNOT00000044866 |
Sf1
|
splicing factor 1 |
| chr2_+_189169877 | 0.01 |
ENSRNOT00000028225
|
She
|
Src homology 2 domain containing E |
| chr19_+_37252843 | 0.01 |
ENSRNOT00000021145
|
E2f4
|
E2F transcription factor 4 |
| chr8_-_48652064 | 0.01 |
ENSRNOT00000030745
ENSRNOT00000083490 |
C2cd2l
|
C2CD2-like |
| chr13_-_60496511 | 0.01 |
ENSRNOT00000004495
|
Cdc73
|
cell division cycle 73 |
| chr10_+_86564928 | 0.01 |
ENSRNOT00000039892
|
Lrrc3c
|
leucine rich repeat containing 3C |
| chr9_+_82053581 | 0.01 |
ENSRNOT00000086375
|
Wnt10a
|
wingless-type MMTV integration site family, member 10A |
| chr9_+_100888501 | 0.01 |
ENSRNOT00000025529
|
Atg4b
|
autophagy related 4B, cysteine peptidase |
| chr10_+_94207336 | 0.01 |
ENSRNOT00000010816
|
Kcnh6
|
potassium voltage-gated channel subfamily H member 6 |
| chr20_-_29511382 | 0.01 |
ENSRNOT00000085026
|
Ddit4
|
DNA-damage-inducible transcript 4 |
| chr19_+_57614628 | 0.01 |
ENSRNOT00000026617
|
Gnpat
|
glyceronephosphate O-acyltransferase |
| chr3_+_172385672 | 0.01 |
ENSRNOT00000090989
|
Gnas
|
GNAS complex locus |
| chr3_+_46286712 | 0.01 |
ENSRNOT00000085563
|
March7
|
membrane associated ring-CH-type finger 7 |
| chr3_+_13838304 | 0.01 |
ENSRNOT00000025067
|
Hspa5
|
heat shock protein family A member 5 |
| chr6_+_136042059 | 0.01 |
ENSRNOT00000088784
|
Mark3
|
microtubule affinity regulating kinase 3 |
| chr4_-_147592699 | 0.01 |
ENSRNOT00000013831
|
Raf1
|
Raf-1 proto-oncogene, serine/threonine kinase |
| chr19_-_57614558 | 0.01 |
ENSRNOT00000025948
|
RGD1562218
|
similar to RIKEN cDNA 0610039J04 |
| chr3_-_52998650 | 0.01 |
ENSRNOT00000009988
|
RGD1560831
|
similar to 40S ribosomal protein S3 |
| chr3_-_170040953 | 0.01 |
ENSRNOT00000005866
|
Cbln4
|
cerebellin 4 precursor |
| chr6_+_93281408 | 0.00 |
ENSRNOT00000009610
|
Frmd6
|
FERM domain containing 6 |
| chr18_+_54206651 | 0.00 |
ENSRNOT00000073295
|
Chsy3
|
chondroitin sulfate synthase 3 |
| chr20_+_6973398 | 0.00 |
ENSRNOT00000041665
|
Fgd2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr7_-_77162148 | 0.00 |
ENSRNOT00000008350
|
Klf10
|
Kruppel-like factor 10 |
| chr16_+_55834458 | 0.00 |
ENSRNOT00000086141
|
AABR07025928.1
|
|
| chr10_-_35800120 | 0.00 |
ENSRNOT00000004361
|
Maml1
|
mastermind-like transcriptional coactivator 1 |
| chr2_-_243224883 | 0.00 |
ENSRNOT00000014139
|
Dapp1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides 1 |
| chr12_-_30501860 | 0.00 |
ENSRNOT00000001227
|
Cct6a
|
chaperonin containing TCP1 subunit 6A |
| chr14_+_86101277 | 0.00 |
ENSRNOT00000018846
|
Aebp1
|
AE binding protein 1 |
| chr3_-_3594475 | 0.00 |
ENSRNOT00000064861
|
RGD1564379
|
RGD1564379 |
| chr3_-_80933283 | 0.00 |
ENSRNOT00000007771
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr1_+_68436917 | 0.00 |
ENSRNOT00000088586
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chr1_-_59347472 | 0.00 |
ENSRNOT00000017718
|
Lnpep
|
leucyl and cystinyl aminopeptidase |
| chr1_+_78933372 | 0.00 |
ENSRNOT00000029810
|
Ccdc8
|
coiled-coil domain containing 8 |
| chr5_-_159293673 | 0.00 |
ENSRNOT00000009081
|
Padi4
|
peptidyl arginine deiminase 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0045575 | basophil activation(GO:0045575) |
| 0.0 | 0.1 | GO:1900623 | regulation of mast cell cytokine production(GO:0032763) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.0 | 0.1 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0014731 | spectrin(GO:0008091) spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0035101 | FACT complex(GO:0035101) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |