Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
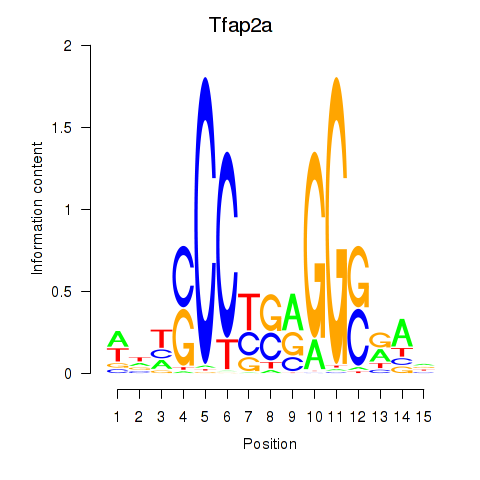
Results for Tfap2a
Z-value: 1.46
Transcription factors associated with Tfap2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2a
|
ENSRNOG00000015522 | transcription factor AP-2 alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2a | rn6_v1_chr17_+_24654902_24654902 | -0.07 | 9.1e-01 | Click! |
Activity profile of Tfap2a motif
Sorted Z-values of Tfap2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_169288871 | 0.51 |
ENSRNOT00000055466
|
Tnfrsf25
|
TNF receptor superfamily member 25 |
| chr7_+_27174882 | 0.50 |
ENSRNOT00000051181
|
Glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr3_+_175426752 | 0.50 |
ENSRNOT00000085718
|
Ss18l1
|
SS18L1, nBAF chromatin remodeling complex subunit |
| chr1_+_266143818 | 0.46 |
ENSRNOT00000026987
|
Sufu
|
SUFU negative regulator of hedgehog signaling |
| chr6_-_65319527 | 0.45 |
ENSRNOT00000005618
|
Stxbp6
|
syntaxin binding protein 6 |
| chr1_-_65681440 | 0.42 |
ENSRNOT00000026305
|
Zfp128
|
zinc finger protein 128 |
| chr1_-_166912524 | 0.41 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr14_-_42221225 | 0.40 |
ENSRNOT00000036103
|
Shisa3
|
shisa family member 3 |
| chr8_+_127144903 | 0.39 |
ENSRNOT00000013530
|
Eomes
|
eomesodermin |
| chr14_+_81819799 | 0.38 |
ENSRNOT00000076840
|
Mxd4
|
Max dimerization protein 4 |
| chr3_+_155160481 | 0.37 |
ENSRNOT00000021133
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr6_+_26399950 | 0.36 |
ENSRNOT00000085424
|
Ift172
|
intraflagellar transport 172 |
| chr19_-_31942180 | 0.36 |
ENSRNOT00000024924
|
Otud4
|
OTU deubiquitinase 4 |
| chr10_-_105368242 | 0.36 |
ENSRNOT00000075293
ENSRNOT00000072230 |
Rnf157
|
ring finger protein 157 |
| chr4_+_56805132 | 0.35 |
ENSRNOT00000010001
ENSRNOT00000085483 |
Irf5
|
interferon regulatory factor 5 |
| chr10_+_58860940 | 0.34 |
ENSRNOT00000056551
ENSRNOT00000074523 |
XAF1
|
XIAP associated factor-1 |
| chr11_+_34051993 | 0.34 |
ENSRNOT00000076473
ENSRNOT00000064751 |
Morc3
|
MORC family CW-type zinc finger 3 |
| chr7_+_142912316 | 0.34 |
ENSRNOT00000010171
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr3_+_108944141 | 0.33 |
ENSRNOT00000034950
|
Fam98b
|
family with sequence similarity 98, member B |
| chr2_+_116028781 | 0.32 |
ENSRNOT00000089477
|
Phc3
|
polyhomeotic homolog 3 |
| chr10_-_85684138 | 0.32 |
ENSRNOT00000017989
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase type 2 beta |
| chr13_-_100740728 | 0.32 |
ENSRNOT00000000074
|
Fbxo28
|
F-box protein 28 |
| chr7_+_142575672 | 0.32 |
ENSRNOT00000080923
ENSRNOT00000008160 |
Scn8a
|
sodium voltage-gated channel alpha subunit 8 |
| chr1_-_260638816 | 0.32 |
ENSRNOT00000065632
ENSRNOT00000017938 ENSRNOT00000077962 |
Pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr14_-_78707861 | 0.32 |
ENSRNOT00000091927
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr7_-_63578490 | 0.32 |
ENSRNOT00000007295
|
Rassf3
|
Ras association domain family member 3 |
| chr1_-_52992213 | 0.31 |
ENSRNOT00000033528
|
Prr18
|
proline rich 18 |
| chr14_-_2032593 | 0.31 |
ENSRNOT00000000037
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr7_+_130474279 | 0.31 |
ENSRNOT00000092388
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr6_+_9483594 | 0.31 |
ENSRNOT00000089272
|
AABR07062799.2
|
|
| chr12_+_2579887 | 0.31 |
ENSRNOT00000068586
ENSRNOT00000043197 |
Prr36
|
proline rich 36 |
| chr17_+_85364483 | 0.31 |
ENSRNOT00000064257
|
Bmi1
|
BMI1 proto-oncogene, polycomb ring finger |
| chr8_+_114866768 | 0.30 |
ENSRNOT00000076731
|
Wdr82
|
WD repeat domain 82 |
| chr1_+_78029555 | 0.30 |
ENSRNOT00000047289
|
Slc8a2
|
solute carrier family 8 member A2 |
| chrX_-_157095274 | 0.29 |
ENSRNOT00000084088
|
Abcd1
|
ATP binding cassette subfamily D member 1 |
| chr1_+_81208212 | 0.29 |
ENSRNOT00000026288
|
Lypd5
|
Ly6/Plaur domain containing 5 |
| chr7_-_145450233 | 0.29 |
ENSRNOT00000092974
ENSRNOT00000021523 |
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr3_-_67668772 | 0.28 |
ENSRNOT00000010247
|
Frzb
|
frizzled-related protein |
| chr20_+_5535432 | 0.27 |
ENSRNOT00000040859
|
Syngap1
|
synaptic Ras GTPase activating protein 1 |
| chr9_+_111028824 | 0.27 |
ENSRNOT00000041418
ENSRNOT00000056457 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr10_+_11786121 | 0.27 |
ENSRNOT00000033919
|
Slx4
|
SLX4 structure-specific endonuclease subunit |
| chr1_-_226887156 | 0.27 |
ENSRNOT00000054809
ENSRNOT00000028347 |
Cd6
|
Cd6 molecule |
| chr17_-_10208360 | 0.27 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr14_+_84150908 | 0.26 |
ENSRNOT00000005516
|
Dusp18
|
dual specificity phosphatase 18 |
| chr6_-_25616995 | 0.26 |
ENSRNOT00000077894
|
Fosl2
|
FOS like 2, AP-1 transcription factor subunit |
| chr13_+_52976507 | 0.26 |
ENSRNOT00000090599
ENSRNOT00000011324 |
Kif21b
|
kinesin family member 21B |
| chr6_+_135610743 | 0.25 |
ENSRNOT00000010906
|
Traf3
|
Tnf receptor-associated factor 3 |
| chr10_+_63829807 | 0.25 |
ENSRNOT00000006407
|
Crk
|
CRK proto-oncogene, adaptor protein |
| chr1_+_199197138 | 0.25 |
ENSRNOT00000065187
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr7_+_11215328 | 0.25 |
ENSRNOT00000061194
|
LOC690617
|
hypothetical protein LOC690617 |
| chr4_+_155653718 | 0.24 |
ENSRNOT00000065419
|
Foxj2
|
forkhead box J2 |
| chr1_-_267245636 | 0.24 |
ENSRNOT00000082799
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr18_-_71701423 | 0.24 |
ENSRNOT00000024716
|
Ctif
|
cap binding complex dependent translation initiation factor |
| chr1_+_212181374 | 0.24 |
ENSRNOT00000085921
|
Adgra1
|
adhesion G protein-coupled receptor A1 |
| chr8_-_22150005 | 0.24 |
ENSRNOT00000041678
|
Tyk2
|
tyrosine kinase 2 |
| chr1_-_163129641 | 0.24 |
ENSRNOT00000083055
|
Capn5
|
calpain 5 |
| chr3_-_170040953 | 0.24 |
ENSRNOT00000005866
|
Cbln4
|
cerebellin 4 precursor |
| chr6_-_132802206 | 0.24 |
ENSRNOT00000080163
ENSRNOT00000050350 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr6_-_111329882 | 0.23 |
ENSRNOT00000074480
|
Ism2
|
isthmin 2 |
| chr5_+_90800082 | 0.23 |
ENSRNOT00000093696
ENSRNOT00000093206 |
Kdm4c
|
lysine demethylase 4C |
| chr1_+_154377447 | 0.23 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr8_-_44327551 | 0.23 |
ENSRNOT00000083939
|
Gramd1b
|
GRAM domain containing 1B |
| chr18_-_77322690 | 0.23 |
ENSRNOT00000058382
|
Nfatc1
|
nuclear factor of activated T-cells 1 |
| chr19_-_25801526 | 0.23 |
ENSRNOT00000003884
|
Nacc1
|
nucleus accumbens associated 1 |
| chr7_+_27309966 | 0.23 |
ENSRNOT00000031871
|
Nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr7_+_127081978 | 0.23 |
ENSRNOT00000022945
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chr1_-_72461547 | 0.23 |
ENSRNOT00000022480
|
Ssc5d
|
scavenger receptor cysteine rich family member with 5 domains |
| chr13_+_110920830 | 0.23 |
ENSRNOT00000077014
ENSRNOT00000076362 |
Kcnh1
|
potassium voltage-gated channel subfamily H member 1 |
| chr11_-_71695000 | 0.23 |
ENSRNOT00000073550
|
Ubxn7
|
UBX domain protein 7 |
| chr2_+_189169877 | 0.23 |
ENSRNOT00000028225
|
She
|
Src homology 2 domain containing E |
| chr15_-_88670349 | 0.23 |
ENSRNOT00000012881
|
Rnf219
|
ring finger protein 219 |
| chr3_+_22640545 | 0.22 |
ENSRNOT00000064507
ENSRNOT00000014452 |
Lhx2
|
LIM homeobox 2 |
| chr5_+_58855773 | 0.22 |
ENSRNOT00000072869
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr9_+_47386626 | 0.22 |
ENSRNOT00000021270
|
Slc9a2
|
solute carrier family 9 member A2 |
| chr2_+_242634399 | 0.22 |
ENSRNOT00000035700
|
Emcn
|
endomucin |
| chr8_+_22189600 | 0.22 |
ENSRNOT00000061100
|
Pde4a
|
phosphodiesterase 4A |
| chr5_+_14415841 | 0.22 |
ENSRNOT00000010682
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr10_-_88060561 | 0.22 |
ENSRNOT00000019133
|
Krt19
|
keratin 19 |
| chr10_-_90030639 | 0.22 |
ENSRNOT00000090456
|
Mpp2
|
membrane palmitoylated protein 2 |
| chr3_-_3855391 | 0.21 |
ENSRNOT00000025764
|
Inpp5e
|
inositol polyphosphate-5-phosphatase E |
| chr12_+_13810180 | 0.21 |
ENSRNOT00000084865
ENSRNOT00000038203 |
Tnrc18
|
trinucleotide repeat containing 18 |
| chr6_+_43234526 | 0.20 |
ENSRNOT00000086808
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr13_-_86671515 | 0.20 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr4_+_78024765 | 0.20 |
ENSRNOT00000043856
|
Krba1
|
KRAB-A domain containing 1 |
| chr12_+_2589818 | 0.20 |
ENSRNOT00000087848
|
AABR07034980.1
|
|
| chr19_+_28263095 | 0.20 |
ENSRNOT00000047230
|
LOC501421
|
hypothetical gene supported by BC082068 |
| chr3_+_120726906 | 0.20 |
ENSRNOT00000051069
|
Bcl2l11
|
BCL2 like 11 |
| chr14_-_72380330 | 0.20 |
ENSRNOT00000082653
ENSRNOT00000081878 ENSRNOT00000006727 |
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr11_+_81972219 | 0.20 |
ENSRNOT00000002452
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr16_+_21061237 | 0.20 |
ENSRNOT00000092136
|
Ncan
|
neurocan |
| chr19_+_37179814 | 0.20 |
ENSRNOT00000085891
ENSRNOT00000020003 |
RGD621098
|
similar to RIKEN cDNA D230025D16Rik |
| chr7_-_77162148 | 0.20 |
ENSRNOT00000008350
|
Klf10
|
Kruppel-like factor 10 |
| chr7_+_80713321 | 0.19 |
ENSRNOT00000091355
ENSRNOT00000078354 |
Oxr1
|
oxidation resistance 1 |
| chr12_-_16813838 | 0.19 |
ENSRNOT00000033551
|
Elfn1
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chr7_-_120672350 | 0.19 |
ENSRNOT00000018126
ENSRNOT00000087800 ENSRNOT00000088424 |
Csnk1e
|
casein kinase 1, epsilon |
| chr4_+_181286455 | 0.19 |
ENSRNOT00000083633
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr5_-_166430254 | 0.19 |
ENSRNOT00000048914
|
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr11_+_61321459 | 0.19 |
ENSRNOT00000002759
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr8_-_111984572 | 0.19 |
ENSRNOT00000014519
|
Tmem108
|
transmembrane protein 108 |
| chr5_-_168123395 | 0.19 |
ENSRNOT00000024932
|
Per3
|
period circadian clock 3 |
| chr6_-_132972511 | 0.19 |
ENSRNOT00000082216
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr2_-_178057157 | 0.18 |
ENSRNOT00000091135
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr2_-_216509629 | 0.18 |
ENSRNOT00000023366
ENSRNOT00000087714 |
Rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr2_+_205783252 | 0.18 |
ENSRNOT00000025633
ENSRNOT00000025600 |
Trim33
|
tripartite motif-containing 33 |
| chr2_+_242882306 | 0.18 |
ENSRNOT00000013661
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr13_-_60496511 | 0.18 |
ENSRNOT00000004495
|
Cdc73
|
cell division cycle 73 |
| chr15_-_52029816 | 0.18 |
ENSRNOT00000013067
|
Slc39a14
|
solute carrier family 39 member 14 |
| chr8_+_61659445 | 0.18 |
ENSRNOT00000023831
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr8_-_56696968 | 0.18 |
ENSRNOT00000016637
|
Zc3h12c
|
zinc finger CCCH type containing 12C |
| chr6_+_122920317 | 0.18 |
ENSRNOT00000068595
ENSRNOT00000082856 |
Ttc8
|
tetratricopeptide repeat domain 8 |
| chr2_-_216509827 | 0.18 |
ENSRNOT00000088533
|
Rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr3_+_109862117 | 0.18 |
ENSRNOT00000083351
ENSRNOT00000070912 |
Thbs1
|
thrombospondin 1 |
| chr12_-_2626310 | 0.18 |
ENSRNOT00000077396
|
Evi5l
|
ecotropic viral integration site 5-like |
| chr2_+_122877286 | 0.17 |
ENSRNOT00000033080
|
Arse
|
arylsulfatase E |
| chr1_-_198900375 | 0.17 |
ENSRNOT00000024969
|
Zfp689
|
zinc finger protein 689 |
| chr5_-_168123030 | 0.17 |
ENSRNOT00000092747
|
Per3
|
period circadian clock 3 |
| chr10_+_94988362 | 0.17 |
ENSRNOT00000066525
|
Cep95
|
centrosomal protein 95 |
| chr19_-_33080998 | 0.17 |
ENSRNOT00000015985
|
Slc10a7
|
solute carrier family 10, member 7 |
| chr5_+_169519212 | 0.17 |
ENSRNOT00000024732
|
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr1_+_78876205 | 0.17 |
ENSRNOT00000022610
|
Pnmal2
|
paraneoplastic Ma antigen family-like 2 |
| chr1_-_225952516 | 0.17 |
ENSRNOT00000043387
|
Incenp
|
inner centromere protein |
| chr10_+_59613398 | 0.17 |
ENSRNOT00000025553
|
Ncbp3
|
nuclear cap binding subunit 3 |
| chr7_-_118474609 | 0.16 |
ENSRNOT00000089076
|
AABR07058464.1
|
|
| chr17_+_10458110 | 0.16 |
ENSRNOT00000089885
|
Rnf44
|
ring finger protein 44 |
| chr8_-_56696523 | 0.16 |
ENSRNOT00000080043
|
Zc3h12c
|
zinc finger CCCH type containing 12C |
| chr5_+_139007642 | 0.16 |
ENSRNOT00000066774
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr2_+_205553163 | 0.16 |
ENSRNOT00000039572
|
Nras
|
neuroblastoma RAS viral oncogene homolog |
| chr12_-_51341663 | 0.16 |
ENSRNOT00000066892
ENSRNOT00000052202 |
Pitpnb
|
phosphatidylinositol transfer protein, beta |
| chr1_-_90520344 | 0.16 |
ENSRNOT00000078598
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr9_+_119517101 | 0.16 |
ENSRNOT00000020476
|
Lpin2
|
lipin 2 |
| chr7_-_11777503 | 0.16 |
ENSRNOT00000026220
|
Amh
|
anti-Mullerian hormone |
| chr7_-_114590119 | 0.16 |
ENSRNOT00000079599
|
Ptk2
|
protein tyrosine kinase 2 |
| chr10_-_56530842 | 0.16 |
ENSRNOT00000077451
|
AABR07029863.3
|
|
| chr7_-_14402010 | 0.16 |
ENSRNOT00000008303
|
Wiz
|
widely-interspaced zinc finger motifs |
| chr5_+_144634143 | 0.16 |
ENSRNOT00000075558
|
RGD1563072
|
similar to hypothetical protein FLJ38984 |
| chr2_+_198823366 | 0.16 |
ENSRNOT00000083769
ENSRNOT00000028814 |
Pias3
|
protein inhibitor of activated STAT, 3 |
| chr3_-_120157866 | 0.15 |
ENSRNOT00000020367
|
LOC100909998
|
zinc finger protein 2-like |
| chr5_+_14415606 | 0.15 |
ENSRNOT00000089273
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr14_+_1462358 | 0.15 |
ENSRNOT00000077243
|
Csf2ra
|
colony stimulating factor 2 receptor alpha subunit |
| chr10_+_89578212 | 0.15 |
ENSRNOT00000028178
|
Arl4d
|
ADP-ribosylation factor like GTPase 4D |
| chr20_+_40618623 | 0.15 |
ENSRNOT00000082043
|
Pkib
|
cAMP-dependent protein kinase inhibitor beta |
| chr8_-_50531423 | 0.15 |
ENSRNOT00000090985
ENSRNOT00000074942 |
Apoc3
|
apolipoprotein C3 |
| chr9_+_70133969 | 0.15 |
ENSRNOT00000040525
|
Adam23
|
ADAM metallopeptidase domain 23 |
| chr5_+_151172206 | 0.15 |
ENSRNOT00000013778
|
Fgr
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr4_-_147893992 | 0.15 |
ENSRNOT00000032158
|
Plxnd1
|
plexin D1 |
| chr1_+_219173002 | 0.15 |
ENSRNOT00000024018
|
Unc93b1
|
unc-93 homolog B1 (C. elegans) |
| chr1_+_183892841 | 0.15 |
ENSRNOT00000015498
|
Pde3b
|
phosphodiesterase 3B |
| chr10_+_54155876 | 0.15 |
ENSRNOT00000072855
|
Gas7
|
growth arrest specific 7 |
| chr1_+_198911911 | 0.15 |
ENSRNOT00000055002
|
Prr14
|
proline rich 14 |
| chr20_-_4380600 | 0.15 |
ENSRNOT00000068712
|
Egfl8
|
EGF-like-domain, multiple 8 |
| chr4_+_7889869 | 0.15 |
ENSRNOT00000014063
|
Pus7
|
pseudouridylate synthase 7 |
| chr12_+_16030788 | 0.15 |
ENSRNOT00000051565
|
Iqce
|
IQ motif containing E |
| chr3_-_153250641 | 0.15 |
ENSRNOT00000008961
|
Samhd1
|
SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 |
| chr10_-_37311625 | 0.15 |
ENSRNOT00000043343
|
Jade2
|
jade family PHD finger 2 |
| chr18_+_14756684 | 0.14 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr7_-_3342491 | 0.14 |
ENSRNOT00000081756
|
Rdh5
|
retinol dehydrogenase 5 |
| chr19_-_10826895 | 0.14 |
ENSRNOT00000090217
|
Rspry1
|
ring finger and SPRY domain containing 1 |
| chr8_+_28352772 | 0.14 |
ENSRNOT00000012391
|
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr1_-_98467262 | 0.14 |
ENSRNOT00000039446
|
Vsig10l
|
V-set and immunoglobulin domain containing 10 like |
| chr18_+_65388685 | 0.14 |
ENSRNOT00000080844
|
Tcf4
|
transcription factor 4 |
| chr13_+_99136871 | 0.14 |
ENSRNOT00000078263
ENSRNOT00000004350 |
Sde2
|
SDE2 telomere maintenance homolog |
| chr20_-_4561062 | 0.14 |
ENSRNOT00000065044
ENSRNOT00000092698 ENSRNOT00000060607 |
Cfb
C2
|
complement factor B complement C2 |
| chr12_-_47793534 | 0.14 |
ENSRNOT00000001588
|
Fam222a
|
family with sequence similarity 222, member A |
| chr5_-_151117042 | 0.14 |
ENSRNOT00000066549
|
Fam76a
|
family with sequence similarity 76, member A |
| chr13_-_81214494 | 0.14 |
ENSRNOT00000004950
ENSRNOT00000082385 |
Prrx1
|
paired related homeobox 1 |
| chr15_-_52288893 | 0.14 |
ENSRNOT00000016209
|
Fam160b2
|
family with sequence similarity 160, member B2 |
| chr20_+_3558827 | 0.14 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr4_-_117575154 | 0.14 |
ENSRNOT00000075813
|
LOC102556148
|
probable N-acetyltransferase CML2-like |
| chr9_+_17086475 | 0.14 |
ENSRNOT00000025660
|
Tjap1
|
tight junction associated protein 1 |
| chr3_-_175644988 | 0.14 |
ENSRNOT00000084452
|
Cables2
|
Cdk5 and Abl enzyme substrate 2 |
| chr5_-_144436509 | 0.14 |
ENSRNOT00000042371
|
Ago3
|
argonaute 3, RISC catalytic component |
| chr4_+_72718458 | 0.14 |
ENSRNOT00000044780
|
Arhgef5
|
Rho guanine nucleotide exchange factor 5 |
| chr6_-_147172022 | 0.14 |
ENSRNOT00000080675
|
Itgb8
|
integrin subunit beta 8 |
| chr7_+_130498199 | 0.14 |
ENSRNOT00000092684
ENSRNOT00000092431 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr1_-_164307084 | 0.14 |
ENSRNOT00000086091
|
Serpinh1
|
serpin family H member 1 |
| chr10_+_72197977 | 0.14 |
ENSRNOT00000003886
|
Myo19
|
myosin XIX |
| chr7_-_31784192 | 0.14 |
ENSRNOT00000010869
|
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr5_+_159967839 | 0.14 |
ENSRNOT00000051317
|
Hspb7
|
heat shock protein family B (small) member 7 |
| chr4_-_67520356 | 0.14 |
ENSRNOT00000014604
|
Braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr10_-_66602987 | 0.13 |
ENSRNOT00000017949
|
Wsb1
|
WD repeat and SOCS box-containing 1 |
| chr1_-_209749204 | 0.13 |
ENSRNOT00000073093
|
LOC100362342
|
rCG47764-like |
| chr18_-_31071371 | 0.13 |
ENSRNOT00000060432
|
Diaph1
|
diaphanous-related formin 1 |
| chr2_+_199831990 | 0.13 |
ENSRNOT00000063783
ENSRNOT00000086588 |
Prkab2
|
protein kinase AMP-activated non-catalytic subunit beta 2 |
| chr8_-_89130991 | 0.13 |
ENSRNOT00000017411
|
Htr1b
|
5-hydroxytryptamine receptor 1B |
| chr1_-_167308827 | 0.13 |
ENSRNOT00000027590
ENSRNOT00000027575 |
Nup98
|
nucleoporin 98 |
| chr1_-_199336451 | 0.13 |
ENSRNOT00000035672
|
Prss53
|
protease, serine, 53 |
| chr7_+_42269784 | 0.13 |
ENSRNOT00000008471
ENSRNOT00000007231 |
Kitlg
|
KIT ligand |
| chr12_-_22126350 | 0.13 |
ENSRNOT00000076328
|
Sap25
|
Sin3A-associated protein 25 |
| chr20_+_4355175 | 0.13 |
ENSRNOT00000000510
|
Gpsm3
|
G-protein signaling modulator 3 |
| chr2_+_95320283 | 0.13 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr6_+_137288810 | 0.13 |
ENSRNOT00000030246
|
Cep170b
|
centrosomal protein 170B |
| chr4_+_157513414 | 0.13 |
ENSRNOT00000078769
|
Pianp
|
PILR alpha associated neural protein |
| chr3_+_172856733 | 0.13 |
ENSRNOT00000009826
|
Edn3
|
endothelin 3 |
| chr7_+_142877086 | 0.13 |
ENSRNOT00000088730
|
Grasp
|
general receptor for phosphoinositides 1 associated scaffold protein |
| chr10_-_104179523 | 0.13 |
ENSRNOT00000005292
|
Slc25a19
|
solute carrier family 25 member 19 |
| chr9_+_50892605 | 0.13 |
ENSRNOT00000033133
|
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr4_-_88649216 | 0.13 |
ENSRNOT00000058626
|
Herc6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr6_-_50786967 | 0.13 |
ENSRNOT00000009566
|
Cbll1
|
Cbl proto-oncogene like 1 |
| chr10_+_52334240 | 0.13 |
ENSRNOT00000005518
|
Zfp18
|
zinc finger protein 18 |
| chr7_+_18440742 | 0.13 |
ENSRNOT00000011513
|
Myo1f
|
myosin IF |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.5 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.3 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.5 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.1 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.3 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.3 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 0.5 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.2 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.3 | GO:1990859 | response to endothelin(GO:1990839) cellular response to endothelin(GO:1990859) |
| 0.1 | 0.3 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.3 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of nitric oxide mediated signal transduction(GO:0010751) regulation of cGMP-mediated signaling(GO:0010752) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.2 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.1 | 0.3 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.1 | 0.3 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.2 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.2 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.4 | GO:0061525 | hindgut development(GO:0061525) |
| 0.1 | 0.2 | GO:0010982 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.2 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.2 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.4 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.4 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.4 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.2 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:1905245 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.1 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.4 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.2 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.4 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.0 | 0.1 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.0 | 0.1 | GO:0072021 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.2 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.0 | 0.1 | GO:1903944 | cholangiocyte apoptotic process(GO:1902488) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.1 | GO:0001552 | ovarian follicle atresia(GO:0001552) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.0 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.3 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.0 | GO:0071038 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.0 | GO:0046356 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.0 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 0.0 | 0.0 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0060059 | skeletal muscle satellite cell differentiation(GO:0014816) embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.0 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.1 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0032242 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.0 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 0.3 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.4 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:1990745 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.0 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.4 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.3 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.3 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.3 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.6 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.2 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.0 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) NADP+ binding(GO:0070401) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.0 | GO:0032142 | single guanine insertion binding(GO:0032142) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |