Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
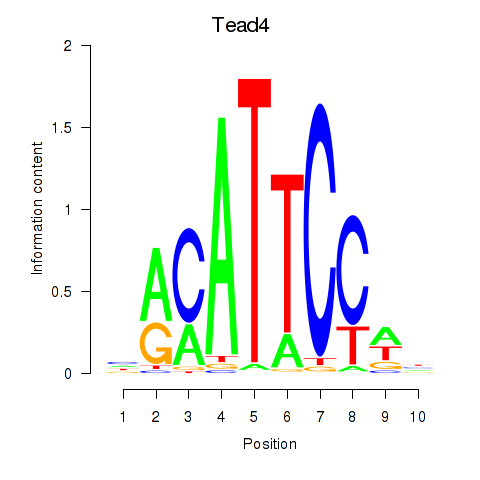
Results for Tead3_Tead4
Z-value: 0.51
Transcription factors associated with Tead3_Tead4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tead3
|
ENSRNOG00000000506 | TEA domain transcription factor 3 |
|
Tead4
|
ENSRNOG00000005608 | TEA domain transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tead3 | rn6_v1_chr20_-_7930929_7930929 | -0.76 | 1.3e-01 | Click! |
| Tead4 | rn6_v1_chr4_-_161587222_161587222 | -0.21 | 7.4e-01 | Click! |
Activity profile of Tead3_Tead4 motif
Sorted Z-values of Tead3_Tead4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_161433410 | 0.43 |
ENSRNOT00000024657
|
Slc12a5
|
solute carrier family 12 member 5 |
| chr20_-_12820466 | 0.21 |
ENSRNOT00000001699
|
Ftcd
|
formimidoyltransferase cyclodeaminase |
| chrX_+_22247088 | 0.18 |
ENSRNOT00000079802
|
Iqsec2
|
IQ motif and Sec7 domain 2 |
| chr3_+_21764377 | 0.18 |
ENSRNOT00000063938
|
Gpr21
|
G protein-coupled receptor 21 |
| chr11_-_64952687 | 0.17 |
ENSRNOT00000087892
|
Popdc2
|
popeye domain containing 2 |
| chr7_-_52404774 | 0.15 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr8_+_48438259 | 0.15 |
ENSRNOT00000059813
|
Mfrp
|
membrane frizzled-related protein |
| chr5_+_74649765 | 0.14 |
ENSRNOT00000075952
|
Palm2
|
paralemmin 2 |
| chr4_+_120672152 | 0.14 |
ENSRNOT00000077231
|
Mgll
|
monoglyceride lipase |
| chr1_+_248402980 | 0.13 |
ENSRNOT00000043517
|
Chchd4
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chrX_+_111122552 | 0.13 |
ENSRNOT00000083566
ENSRNOT00000085078 ENSRNOT00000090928 |
Cldn2
|
claudin 2 |
| chr10_-_62699723 | 0.12 |
ENSRNOT00000086706
|
Coro6
|
coronin 6 |
| chr4_+_181286455 | 0.12 |
ENSRNOT00000083633
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr17_+_86408151 | 0.11 |
ENSRNOT00000022734
|
Otud1
|
OTU deubiquitinase 1 |
| chr15_-_76789298 | 0.11 |
ENSRNOT00000077853
|
AABR07018857.1
|
|
| chr8_-_55162421 | 0.11 |
ENSRNOT00000083102
|
Dixdc1
|
DIX domain containing 1 |
| chr8_-_116361343 | 0.11 |
ENSRNOT00000066296
|
Sema3b
|
semaphorin 3B |
| chr10_-_13374808 | 0.11 |
ENSRNOT00000060888
|
Prss22
|
protease, serine, 22 |
| chr18_+_55797198 | 0.11 |
ENSRNOT00000026334
ENSRNOT00000026394 |
Dctn4
|
dynactin subunit 4 |
| chr1_-_78521999 | 0.10 |
ENSRNOT00000021223
|
Arhgap35
|
Rho GTPase activating protein 35 |
| chr6_+_135513650 | 0.10 |
ENSRNOT00000010676
|
Rcor1
|
REST corepressor 1 |
| chr9_-_45206427 | 0.10 |
ENSRNOT00000042227
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr1_-_204817080 | 0.10 |
ENSRNOT00000077956
|
Fam53b
|
family with sequence similarity 53, member B |
| chr12_-_37185548 | 0.10 |
ENSRNOT00000001338
|
LOC102556092
|
uncharacterized LOC102556092 |
| chr2_+_150756185 | 0.10 |
ENSRNOT00000088461
ENSRNOT00000036808 |
Mbnl1
|
muscleblind-like splicing regulator 1 |
| chr8_-_104155775 | 0.09 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chr7_+_58419197 | 0.09 |
ENSRNOT00000085829
|
Zfc3h1
|
zinc finger, C3H1-type containing |
| chr13_+_74154835 | 0.09 |
ENSRNOT00000059524
|
Abl2
|
ABL proto-oncogene 2, non-receptor tyrosine kinase |
| chr1_+_84009268 | 0.09 |
ENSRNOT00000057230
ENSRNOT00000081121 |
RGD1560854
|
similar to FLJ41131 protein |
| chr8_+_48437918 | 0.09 |
ENSRNOT00000085578
|
Mfrp
|
membrane frizzled-related protein |
| chr3_+_51737904 | 0.09 |
ENSRNOT00000007069
|
Scn2a
|
sodium voltage-gated channel alpha subunit 2 |
| chr1_-_43638161 | 0.09 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr5_+_5616483 | 0.08 |
ENSRNOT00000011026
|
Ncoa2
|
nuclear receptor coactivator 2 |
| chr9_+_15582564 | 0.08 |
ENSRNOT00000020640
|
LOC100912849
|
uncharacterized LOC100912849 |
| chr2_-_187820952 | 0.08 |
ENSRNOT00000092762
|
Sema4a
|
semaphorin 4A |
| chr20_+_3558827 | 0.08 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr6_-_99783047 | 0.07 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr4_+_152892388 | 0.07 |
ENSRNOT00000056198
ENSRNOT00000090218 ENSRNOT00000075895 |
Kdm5a
|
lysine demethylase 5A |
| chr2_-_116413043 | 0.07 |
ENSRNOT00000078148
|
Mynn
|
myoneurin |
| chr5_+_75392790 | 0.07 |
ENSRNOT00000048457
|
Musk
|
muscle associated receptor tyrosine kinase |
| chr19_+_43163129 | 0.07 |
ENSRNOT00000073721
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr11_+_15436269 | 0.07 |
ENSRNOT00000002150
ENSRNOT00000084928 |
Usp25
|
ubiquitin specific peptidase 25 |
| chr11_-_82664554 | 0.07 |
ENSRNOT00000002425
ENSRNOT00000087105 |
Senp2
|
Sumo1/sentrin/SMT3 specific peptidase 2 |
| chr9_+_16003058 | 0.07 |
ENSRNOT00000081621
ENSRNOT00000021158 |
Ubr2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr8_-_84632817 | 0.07 |
ENSRNOT00000076942
|
Mlip
|
muscular LMNA-interacting protein |
| chr18_+_57286322 | 0.07 |
ENSRNOT00000026174
|
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chrX_+_70596901 | 0.06 |
ENSRNOT00000088114
|
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr9_+_54212767 | 0.06 |
ENSRNOT00000078073
ENSRNOT00000090026 |
Gls
|
glutaminase |
| chr6_-_26624092 | 0.06 |
ENSRNOT00000008113
|
Trim54
|
tripartite motif-containing 54 |
| chr3_-_56030744 | 0.06 |
ENSRNOT00000089637
|
Ccdc173
|
coiled-coil domain containing 173 |
| chr9_-_55256340 | 0.06 |
ENSRNOT00000028907
|
Sdpr
|
serum deprivation response |
| chr3_+_6430589 | 0.06 |
ENSRNOT00000012334
|
Col5a1
|
collagen type V alpha 1 chain |
| chr1_-_18058055 | 0.06 |
ENSRNOT00000020988
|
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr7_-_115045802 | 0.06 |
ENSRNOT00000040236
ENSRNOT00000071818 |
Mroh5
|
maestro heat-like repeat family member 5 |
| chr15_+_86243148 | 0.06 |
ENSRNOT00000084471
ENSRNOT00000090727 |
Lmo7
|
LIM domain 7 |
| chr10_-_39373437 | 0.06 |
ENSRNOT00000058907
|
Slc22a5
|
solute carrier family 22 member 5 |
| chr2_+_127489771 | 0.06 |
ENSRNOT00000093581
|
Intu
|
inturned planar cell polarity protein |
| chr6_-_126710854 | 0.06 |
ENSRNOT00000081127
ENSRNOT00000064914 |
Btbd7
|
BTB domain containing 7 |
| chr1_-_100530183 | 0.06 |
ENSRNOT00000067754
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr2_+_195996521 | 0.06 |
ENSRNOT00000082849
|
Pogz
|
pogo transposable element with ZNF domain |
| chr9_-_52238564 | 0.05 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr10_+_48995521 | 0.05 |
ENSRNOT00000072548
|
AABR07029742.2
|
|
| chr7_-_144319804 | 0.05 |
ENSRNOT00000070855
|
LOC100912282
|
calcium-binding and coiled-coil domain-containing protein 1-like |
| chr5_+_164796185 | 0.05 |
ENSRNOT00000010779
|
Nppb
|
natriuretic peptide B |
| chr2_+_3662763 | 0.05 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr9_+_88357556 | 0.05 |
ENSRNOT00000020669
|
Col4a3
|
collagen type IV alpha 3 chain |
| chr4_+_6282278 | 0.05 |
ENSRNOT00000010349
|
Kmt2c
|
lysine methyltransferase 2C |
| chr7_+_130532435 | 0.05 |
ENSRNOT00000092672
ENSRNOT00000092535 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr18_-_68551558 | 0.05 |
ENSRNOT00000016369
|
Rab27b
|
RAB27B, member RAS oncogene family |
| chr3_+_8537350 | 0.05 |
ENSRNOT00000079640
|
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr14_+_79205466 | 0.05 |
ENSRNOT00000085534
|
Tbc1d14
|
TBC1 domain family, member 14 |
| chrX_-_26376467 | 0.05 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr2_-_165739874 | 0.05 |
ENSRNOT00000014384
|
Kpna4
|
karyopherin subunit alpha 4 |
| chr13_-_70625842 | 0.05 |
ENSRNOT00000092499
|
Lamc2
|
laminin subunit gamma 2 |
| chr19_+_43163562 | 0.05 |
ENSRNOT00000048900
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr1_+_46835971 | 0.05 |
ENSRNOT00000066798
|
Synj2
|
synaptojanin 2 |
| chr7_-_82687130 | 0.05 |
ENSRNOT00000006791
|
Tmem74
|
transmembrane protein 74 |
| chr1_+_44311513 | 0.05 |
ENSRNOT00000065386
|
Tiam2
|
T-cell lymphoma invasion and metastasis 2 |
| chr10_+_74959285 | 0.05 |
ENSRNOT00000010296
|
Rnf43
|
ring finger protein 43 |
| chr8_+_82878941 | 0.05 |
ENSRNOT00000014846
|
Bmp5
|
bone morphogenetic protein 5 |
| chr9_+_73334618 | 0.05 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr5_-_83648044 | 0.04 |
ENSRNOT00000046725
|
RGD1561195
|
similar to ribosomal protein L31 |
| chr1_+_85386470 | 0.04 |
ENSRNOT00000093332
ENSRNOT00000044326 |
Plekhg2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr3_+_111825162 | 0.04 |
ENSRNOT00000066685
|
Pla2g4b
|
phospholipase A2 group IVB |
| chr1_+_15642153 | 0.04 |
ENSRNOT00000079845
|
Map7
|
microtubule-associated protein 7 |
| chr6_+_73345392 | 0.04 |
ENSRNOT00000088378
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr20_+_46428124 | 0.04 |
ENSRNOT00000000327
|
Foxo3
|
forkhead box O3 |
| chr6_-_110681408 | 0.04 |
ENSRNOT00000078265
|
Angel1
|
angel homolog 1 |
| chr9_-_94495333 | 0.04 |
ENSRNOT00000021507
|
Kcnj13
|
potassium voltage-gated channel subfamily J member 13 |
| chr2_-_219458271 | 0.04 |
ENSRNOT00000064735
|
Cdc14a
|
cell division cycle 14A |
| chr10_-_88874528 | 0.04 |
ENSRNOT00000026783
|
Ptrf
|
polymerase I and transcript release factor |
| chr3_-_176926722 | 0.04 |
ENSRNOT00000020168
|
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr17_+_53231343 | 0.04 |
ENSRNOT00000021703
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chrX_+_51286737 | 0.04 |
ENSRNOT00000035692
|
Dmd
|
dystrophin |
| chr19_+_54766589 | 0.04 |
ENSRNOT00000025894
|
Banp
|
Btg3 associated nuclear protein |
| chr9_-_80166807 | 0.04 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr5_+_169274785 | 0.04 |
ENSRNOT00000082586
|
Plekhg5
|
pleckstrin homology and RhoGEF domain containing G5 |
| chr4_+_56711275 | 0.04 |
ENSRNOT00000088149
|
Flnc
|
filamin C |
| chr14_-_39112600 | 0.04 |
ENSRNOT00000003170
|
Gabrb1
|
gamma-aminobutyric acid type A receptor beta 1 subunit |
| chr10_+_92628356 | 0.04 |
ENSRNOT00000072480
|
Myl4
|
myosin, light chain 4 |
| chr4_-_64981384 | 0.04 |
ENSRNOT00000017338
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr15_-_8914501 | 0.04 |
ENSRNOT00000008752
|
Thrb
|
thyroid hormone receptor beta |
| chr11_-_31238026 | 0.04 |
ENSRNOT00000032366
|
RGD1562726
|
similar to Putative protein C21orf62 homolog |
| chr1_+_266053002 | 0.04 |
ENSRNOT00000026235
|
Nfkb2
|
nuclear factor kappa B subunit 2 |
| chr3_+_103753238 | 0.04 |
ENSRNOT00000007144
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr7_-_80457816 | 0.04 |
ENSRNOT00000039430
|
AABR07057617.1
|
|
| chr3_-_94767239 | 0.04 |
ENSRNOT00000056808
|
Qser1
|
glutamine and serine rich 1 |
| chr2_+_182006242 | 0.04 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr6_-_80102635 | 0.03 |
ENSRNOT00000084611
|
Sec23a
|
Sec23 homolog A, coat complex II component |
| chr8_+_22937916 | 0.03 |
ENSRNOT00000088255
ENSRNOT00000049993 |
Ccdc159
|
coiled-coil domain containing 159 |
| chr7_-_81592206 | 0.03 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr5_-_59085676 | 0.03 |
ENSRNOT00000068105
|
Msmp
|
microseminoprotein, prostate associated |
| chr9_+_47281961 | 0.03 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chr17_-_84247038 | 0.03 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr8_-_80631873 | 0.03 |
ENSRNOT00000091661
ENSRNOT00000080662 |
Unc13c
|
unc-13 homolog C |
| chr2_-_52531083 | 0.03 |
ENSRNOT00000089303
ENSRNOT00000021608 |
Zfp131
|
zinc finger protein 131 |
| chr1_-_250774105 | 0.03 |
ENSRNOT00000079227
|
Sgms1
|
sphingomyelin synthase 1 |
| chr4_-_56114254 | 0.03 |
ENSRNOT00000010673
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr3_+_51883559 | 0.03 |
ENSRNOT00000007197
|
Csrnp3
|
cysteine and serine rich nuclear protein 3 |
| chr5_+_27326762 | 0.03 |
ENSRNOT00000066191
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr3_-_170458676 | 0.03 |
ENSRNOT00000035071
|
Gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr4_+_183896303 | 0.03 |
ENSRNOT00000055437
|
RGD1309621
|
similar to hypothetical protein FLJ10652 |
| chr14_+_37551940 | 0.03 |
ENSRNOT00000003061
|
Fryl
|
FRY like transcription coactivator |
| chr12_+_40895515 | 0.03 |
ENSRNOT00000046323
|
Ptpn11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr1_+_88078350 | 0.03 |
ENSRNOT00000048677
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr1_+_224998172 | 0.03 |
ENSRNOT00000026321
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr1_-_174289647 | 0.03 |
ENSRNOT00000064902
|
St5
|
suppression of tumorigenicity 5 |
| chr5_-_120083904 | 0.03 |
ENSRNOT00000064542
|
Jak1
|
Janus kinase 1 |
| chr6_-_136436620 | 0.03 |
ENSRNOT00000067118
|
Ppp1r13b
|
protein phosphatase 1, regulatory subunit 13B |
| chr9_+_46996156 | 0.03 |
ENSRNOT00000019673
|
Il1r1
|
interleukin 1 receptor type 1 |
| chr1_-_67302751 | 0.03 |
ENSRNOT00000041518
|
Vom1r42
|
vomeronasal 1 receptor 42 |
| chr13_+_98311827 | 0.03 |
ENSRNOT00000082844
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr6_-_136436818 | 0.03 |
ENSRNOT00000082600
|
Ppp1r13b
|
protein phosphatase 1, regulatory subunit 13B |
| chr15_+_105640097 | 0.03 |
ENSRNOT00000014300
|
Mbnl2
|
muscleblind-like splicing regulator 2 |
| chr5_-_152458023 | 0.03 |
ENSRNOT00000031225
|
Cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr5_+_151362019 | 0.02 |
ENSRNOT00000077083
ENSRNOT00000067801 |
Wasf2
|
WAS protein family, member 2 |
| chr16_+_71738718 | 0.02 |
ENSRNOT00000022097
|
Plekha2
|
pleckstrin homology domain containing A2 |
| chr14_-_6645257 | 0.02 |
ENSRNOT00000002916
|
Pkd2
|
polycystin 2, transient receptor potential cation channel |
| chr7_-_98197087 | 0.02 |
ENSRNOT00000010484
ENSRNOT00000079961 |
Klhl38
|
kelch-like family member 38 |
| chr20_+_2194709 | 0.02 |
ENSRNOT00000001017
|
Trim15
|
tripartite motif containing 15 |
| chr14_-_114692764 | 0.02 |
ENSRNOT00000008210
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr17_-_76281660 | 0.02 |
ENSRNOT00000058070
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr19_-_55367353 | 0.02 |
ENSRNOT00000091139
|
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr1_+_60106034 | 0.02 |
ENSRNOT00000014930
|
AC091336.1
|
|
| chr10_-_104628676 | 0.02 |
ENSRNOT00000010466
|
Unc13d
|
unc-13 homolog D |
| chr10_-_13580821 | 0.02 |
ENSRNOT00000009735
|
Ntn3
|
netrin 3 |
| chr4_+_70572942 | 0.02 |
ENSRNOT00000051964
|
AC142181.1
|
|
| chr10_+_63778755 | 0.02 |
ENSRNOT00000090481
|
Inpp5k
|
inositol polyphosphate-5-phosphatase K |
| chr4_-_17594598 | 0.02 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr12_+_8085525 | 0.02 |
ENSRNOT00000089804
ENSRNOT00000001234 |
Slc7a1
|
solute carrier family 7 member 1 |
| chr5_-_68139199 | 0.02 |
ENSRNOT00000009987
|
Plppr1
|
phospholipid phosphatase related 1 |
| chr5_+_113725717 | 0.02 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chr1_-_204322544 | 0.02 |
ENSRNOT00000022157
|
Chst15
|
carbohydrate sulfotransferase 15 |
| chrX_+_70596576 | 0.02 |
ENSRNOT00000045082
ENSRNOT00000003741 ENSRNOT00000076079 |
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr9_+_16737642 | 0.02 |
ENSRNOT00000061430
|
Srf
|
serum response factor |
| chr7_-_75098331 | 0.02 |
ENSRNOT00000013344
|
Rnf19a
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr3_-_38741113 | 0.02 |
ENSRNOT00000006549
|
Prpf40a
|
pre-mRNA processing factor 40 homolog A |
| chr13_+_88265331 | 0.02 |
ENSRNOT00000031190
|
Ccdc190
|
coiled-coil domain containing 190 |
| chr15_-_51168384 | 0.02 |
ENSRNOT00000021636
|
Slc25a37
|
solute carrier family 25 member 37 |
| chr15_+_33755641 | 0.02 |
ENSRNOT00000087430
|
Thtpa
|
thiamine triphosphatase |
| chr3_+_79729739 | 0.02 |
ENSRNOT00000084833
|
Kbtbd4
|
kelch repeat and BTB domain containing 4 |
| chr10_-_68926264 | 0.02 |
ENSRNOT00000008661
|
Phb-ps1
|
prohibitin, pseudogene 1 |
| chr15_+_4064706 | 0.02 |
ENSRNOT00000011956
|
Synpo2l
|
synaptopodin 2-like |
| chr7_-_34951644 | 0.02 |
ENSRNOT00000030015
|
Vezt
|
vezatin, adherens junctions transmembrane protein |
| chr8_+_48406260 | 0.02 |
ENSRNOT00000009975
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr3_-_151548995 | 0.02 |
ENSRNOT00000071825
|
LOC102550306
|
uncharacterized LOC102550306 |
| chrX_+_77119911 | 0.02 |
ENSRNOT00000080141
|
Atp7a
|
ATPase copper transporting alpha |
| chr10_-_90657185 | 0.02 |
ENSRNOT00000003685
|
Ccdc43
|
coiled-coil domain containing 43 |
| chr3_-_105214989 | 0.02 |
ENSRNOT00000037895
|
Grem1
|
gremlin 1, DAN family BMP antagonist |
| chr6_-_21135880 | 0.02 |
ENSRNOT00000051239
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chrX_-_135419704 | 0.02 |
ENSRNOT00000044435
|
Zfp280c
|
zinc finger protein 280C |
| chr7_+_34952011 | 0.02 |
ENSRNOT00000077666
|
Fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr4_+_110699557 | 0.02 |
ENSRNOT00000030588
ENSRNOT00000092261 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr7_+_3216497 | 0.02 |
ENSRNOT00000008909
|
Mmp19
|
matrix metallopeptidase 19 |
| chr2_-_41784929 | 0.02 |
ENSRNOT00000086851
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr5_-_135285311 | 0.01 |
ENSRNOT00000088261
ENSRNOT00000085235 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr10_-_72545141 | 0.01 |
ENSRNOT00000033684
|
Appbp2
|
amyloid beta precursor protein binding protein 2 |
| chr5_-_158439078 | 0.01 |
ENSRNOT00000025517
|
Klhdc7a
|
kelch domain containing 7A |
| chr11_-_38351184 | 0.01 |
ENSRNOT00000046318
|
Prdm15
|
PR/SET domain 15 |
| chr1_-_31055453 | 0.01 |
ENSRNOT00000031083
|
Soga3
|
SOGA family member 3 |
| chr7_-_122644054 | 0.01 |
ENSRNOT00000025941
|
Dnajb7
|
DnaJ heat shock protein family (Hsp40) member B7 |
| chr3_+_52588288 | 0.01 |
ENSRNOT00000081962
|
AC122968.1
|
|
| chr20_-_47306318 | 0.01 |
ENSRNOT00000075151
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr3_+_152909189 | 0.01 |
ENSRNOT00000066341
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr3_+_79860179 | 0.01 |
ENSRNOT00000081160
ENSRNOT00000068444 |
Rapsn
|
receptor-associated protein of the synapse |
| chr1_-_217720221 | 0.01 |
ENSRNOT00000039677
ENSRNOT00000092939 |
Ppfia1
|
PTPRF interacting protein alpha 1 |
| chrX_-_138972684 | 0.01 |
ENSRNOT00000040165
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr17_+_82065937 | 0.01 |
ENSRNOT00000051349
|
Arl5b
|
ADP-ribosylation factor like GTPase 5B |
| chr7_-_92882068 | 0.01 |
ENSRNOT00000037809
|
Ext1
|
exostosin glycosyltransferase 1 |
| chr4_-_66336901 | 0.01 |
ENSRNOT00000035326
|
AABR07060293.1
|
|
| chr11_+_27208564 | 0.01 |
ENSRNOT00000002158
|
Map3k7cl
|
MAP3K7 C-terminal like |
| chr10_+_59000397 | 0.01 |
ENSRNOT00000021134
|
Mybbp1a
|
MYB binding protein 1a |
| chr3_+_122816924 | 0.01 |
ENSRNOT00000045604
|
RGD1561317
|
similar to ribosomal protein L31 |
| chr5_+_171478555 | 0.01 |
ENSRNOT00000091417
ENSRNOT00000000169 |
LOC100911486
|
multiple epidermal growth factor-like domains protein 6-like |
| chr4_+_29978739 | 0.01 |
ENSRNOT00000011756
|
Ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr13_-_107886476 | 0.01 |
ENSRNOT00000077282
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr4_+_31333970 | 0.01 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr3_+_45277348 | 0.01 |
ENSRNOT00000059246
|
Pkp4
|
plakophilin 4 |
| chr3_-_23474170 | 0.01 |
ENSRNOT00000039410
|
Scai
|
suppressor of cancer cell invasion |
| chr7_-_140546908 | 0.01 |
ENSRNOT00000077502
|
Kmt2d
|
lysine methyltransferase 2D |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tead3_Tead4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 0.1 | 0.2 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:1901726 | negative regulation of histone deacetylase activity(GO:1901726) |
| 0.0 | 0.1 | GO:0045887 | positive regulation of synaptic growth at neuromuscular junction(GO:0045887) regulation of protein geranylgeranylation(GO:2000539) positive regulation of protein geranylgeranylation(GO:2000541) |
| 0.0 | 0.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) antibiotic transport(GO:0042891) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.0 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.0 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.0 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.1 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |