Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
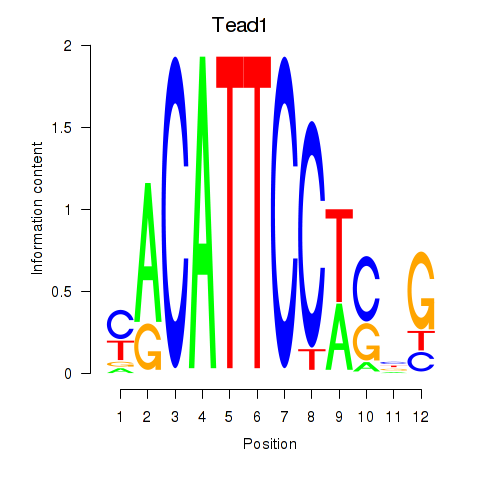
Results for Tead1
Z-value: 0.17
Transcription factors associated with Tead1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tead1
|
ENSRNOG00000015488 | TEA domain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tead1 | rn6_v1_chr1_+_177569618_177569618 | -0.27 | 6.6e-01 | Click! |
Activity profile of Tead1 motif
Sorted Z-values of Tead1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_86408151 | 0.06 |
ENSRNOT00000022734
|
Otud1
|
OTU deubiquitinase 1 |
| chr4_+_181286455 | 0.05 |
ENSRNOT00000083633
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr6_+_109617355 | 0.04 |
ENSRNOT00000011599
|
Flvcr2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr10_+_104523996 | 0.03 |
ENSRNOT00000065339
ENSRNOT00000086747 |
Itgb4
|
integrin subunit beta 4 |
| chr7_-_52404774 | 0.03 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr13_-_86671515 | 0.03 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr20_-_12820466 | 0.03 |
ENSRNOT00000001699
|
Ftcd
|
formimidoyltransferase cyclodeaminase |
| chr3_-_23474170 | 0.03 |
ENSRNOT00000039410
|
Scai
|
suppressor of cancer cell invasion |
| chr1_+_214203861 | 0.03 |
ENSRNOT00000023219
|
Rassf7
|
Ras association domain family member 7 |
| chr2_-_140618405 | 0.03 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr5_-_158439078 | 0.03 |
ENSRNOT00000025517
|
Klhdc7a
|
kelch domain containing 7A |
| chr16_-_49820235 | 0.03 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr19_+_54766589 | 0.03 |
ENSRNOT00000025894
|
Banp
|
Btg3 associated nuclear protein |
| chr4_+_152892388 | 0.03 |
ENSRNOT00000056198
ENSRNOT00000090218 ENSRNOT00000075895 |
Kdm5a
|
lysine demethylase 5A |
| chr5_+_59080765 | 0.03 |
ENSRNOT00000021888
ENSRNOT00000064419 |
Rgp1
|
RGP1 homolog, RAB6A GEF complex partner 1 |
| chr10_-_104482838 | 0.02 |
ENSRNOT00000007246
|
Recql5
|
RecQ like helicase 5 |
| chr1_-_131460473 | 0.02 |
ENSRNOT00000084336
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr4_-_64981384 | 0.02 |
ENSRNOT00000017338
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr19_-_11341863 | 0.02 |
ENSRNOT00000025694
|
Mt4
|
metallothionein 4 |
| chr1_-_18058055 | 0.02 |
ENSRNOT00000020988
|
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr10_-_62699723 | 0.02 |
ENSRNOT00000086706
|
Coro6
|
coronin 6 |
| chr1_+_192613372 | 0.02 |
ENSRNOT00000016632
|
Cacng3
|
calcium voltage-gated channel auxiliary subunit gamma 3 |
| chr6_-_125060482 | 0.02 |
ENSRNOT00000034961
|
Ppp4r3a
|
protein phosphatase 4, regulatory subunit 3A |
| chr11_+_64522130 | 0.02 |
ENSRNOT00000004315
|
Upk1b
|
uroplakin 1B |
| chr19_+_43163129 | 0.02 |
ENSRNOT00000073721
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr5_+_75392790 | 0.02 |
ENSRNOT00000048457
|
Musk
|
muscle associated receptor tyrosine kinase |
| chr10_-_88874528 | 0.02 |
ENSRNOT00000026783
|
Ptrf
|
polymerase I and transcript release factor |
| chr17_-_10004321 | 0.02 |
ENSRNOT00000042394
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr13_+_52662996 | 0.02 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr7_-_140437467 | 0.01 |
ENSRNOT00000087181
|
Fkbp11
|
FK506 binding protein 11 |
| chr9_-_52238564 | 0.01 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr4_+_152892599 | 0.01 |
ENSRNOT00000079090
|
Kdm5a
|
lysine demethylase 5A |
| chr3_+_161433410 | 0.01 |
ENSRNOT00000024657
|
Slc12a5
|
solute carrier family 12 member 5 |
| chr11_-_60679555 | 0.01 |
ENSRNOT00000059735
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr4_-_178168690 | 0.01 |
ENSRNOT00000020729
|
Sox5
|
SRY box 5 |
| chr11_-_38351184 | 0.01 |
ENSRNOT00000046318
|
Prdm15
|
PR/SET domain 15 |
| chr13_+_68709243 | 0.01 |
ENSRNOT00000066419
|
Ivns1abp
|
influenza virus NS1A binding protein |
| chr6_-_76079283 | 0.01 |
ENSRNOT00000082198
|
Ppp2r3c
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr1_+_101517714 | 0.01 |
ENSRNOT00000028423
|
Plekha4
|
pleckstrin homology domain containing A4 |
| chr8_-_73194837 | 0.01 |
ENSRNOT00000024885
ENSRNOT00000081450 ENSRNOT00000064613 |
Tln2
|
talin 2 |
| chr1_-_261371508 | 0.01 |
ENSRNOT00000019978
|
Avpi1
|
arginine vasopressin-induced 1 |
| chr4_+_120672152 | 0.01 |
ENSRNOT00000077231
|
Mgll
|
monoglyceride lipase |
| chr6_+_109617596 | 0.01 |
ENSRNOT00000085031
ENSRNOT00000089972 ENSRNOT00000082402 |
Flvcr2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr2_+_235596123 | 0.01 |
ENSRNOT00000074816
|
Col25a1
|
collagen type XXV alpha 1 chain |
| chr3_+_79860179 | 0.01 |
ENSRNOT00000081160
ENSRNOT00000068444 |
Rapsn
|
receptor-associated protein of the synapse |
| chr1_+_46835971 | 0.01 |
ENSRNOT00000066798
|
Synj2
|
synaptojanin 2 |
| chr10_-_70646495 | 0.01 |
ENSRNOT00000071218
|
Gas2l2
|
growth arrest-specific 2 like 2 |
| chr19_+_43163562 | 0.01 |
ENSRNOT00000048900
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr1_-_216703534 | 0.01 |
ENSRNOT00000027972
|
Phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chrX_+_77119911 | 0.01 |
ENSRNOT00000080141
|
Atp7a
|
ATPase copper transporting alpha |
| chr18_+_57286322 | 0.01 |
ENSRNOT00000026174
|
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr2_-_189333015 | 0.01 |
ENSRNOT00000076424
ENSRNOT00000076952 |
Hax1
|
HCLS1 associated protein X-1 |
| chr10_-_54323439 | 0.01 |
ENSRNOT00000004946
|
Glp2r
|
glucagon-like peptide 2 receptor |
| chr13_+_74154835 | 0.01 |
ENSRNOT00000059524
|
Abl2
|
ABL proto-oncogene 2, non-receptor tyrosine kinase |
| chr7_-_75098331 | 0.00 |
ENSRNOT00000013344
|
Rnf19a
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr17_-_62462644 | 0.00 |
ENSRNOT00000024545
|
Ccny
|
cyclin Y |
| chr18_+_55797198 | 0.00 |
ENSRNOT00000026334
ENSRNOT00000026394 |
Dctn4
|
dynactin subunit 4 |
| chr1_-_88908750 | 0.00 |
ENSRNOT00000028297
|
Aplp1
|
amyloid beta precursor like protein 1 |
| chr9_-_94495333 | 0.00 |
ENSRNOT00000021507
|
Kcnj13
|
potassium voltage-gated channel subfamily J member 13 |
| chr3_+_80362858 | 0.00 |
ENSRNOT00000021353
ENSRNOT00000086242 |
Lrp4
|
LDL receptor related protein 4 |
| chr1_-_100530183 | 0.00 |
ENSRNOT00000067754
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr10_+_65591622 | 0.00 |
ENSRNOT00000015207
|
Pigs
|
phosphatidylinositol glycan anchor biosynthesis, class S |
| chr5_+_164796185 | 0.00 |
ENSRNOT00000010779
|
Nppb
|
natriuretic peptide B |
| chr15_-_41890716 | 0.00 |
ENSRNOT00000020419
|
Spryd7
|
SPRY domain containing 7 |
| chr3_-_79390956 | 0.00 |
ENSRNOT00000077943
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr10_-_85636902 | 0.00 |
ENSRNOT00000017241
|
Pcgf2
|
polycomb group ring finger 2 |
| chr12_+_48316143 | 0.00 |
ENSRNOT00000084511
|
Svop
|
SV2 related protein |
| chr8_+_111210811 | 0.00 |
ENSRNOT00000011347
|
Amotl2
|
angiomotin like 2 |
| chr8_-_61595032 | 0.00 |
ENSRNOT00000023428
|
Snx33
|
sorting nexin 33 |
| chr13_+_26172243 | 0.00 |
ENSRNOT00000003840
|
Phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr6_-_122721496 | 0.00 |
ENSRNOT00000079697
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr9_-_55256340 | 0.00 |
ENSRNOT00000028907
|
Sdpr
|
serum deprivation response |
| chr5_+_28850950 | 0.00 |
ENSRNOT00000009759
|
Tmem64
|
transmembrane protein 64 |
| chr9_+_15166118 | 0.00 |
ENSRNOT00000020432
|
Mdfi
|
MyoD family inhibitor |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tead1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:1901726 | negative regulation of histone deacetylase activity(GO:1901726) |
| 0.0 | 0.0 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |