Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Tcf7_Tcf7l2
Z-value: 1.06
Transcription factors associated with Tcf7_Tcf7l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
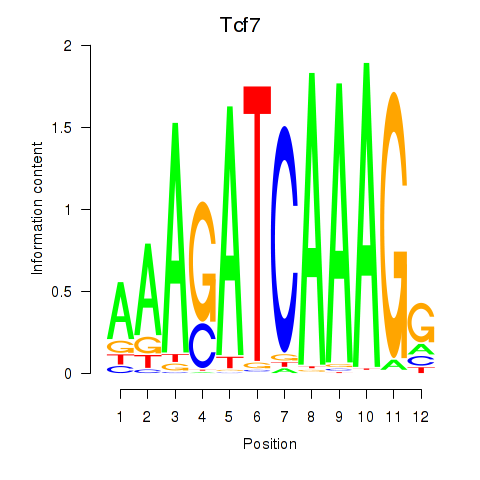
Tcf7
|
ENSRNOG00000005872 | transcription factor 7 (T-cell specific, HMG-box) |
|
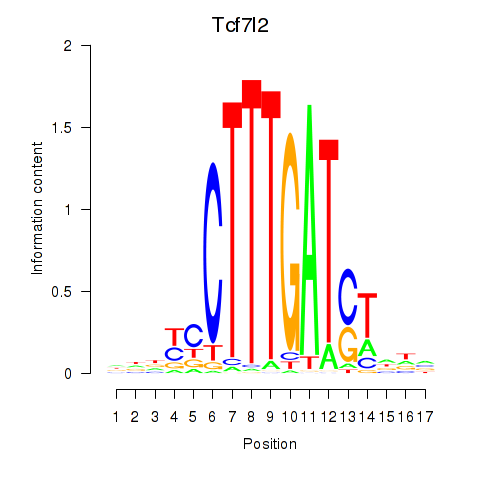
Tcf7l2
|
ENSRNOG00000049232 | transcription factor 7 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf7 | rn6_v1_chr10_-_37645802_37645802 | 0.23 | 7.1e-01 | Click! |
| AABR07007000.1 | rn6_v1_chr1_-_276012351_276012351 | -0.14 | 8.2e-01 | Click! |
Activity profile of Tcf7_Tcf7l2 motif
Sorted Z-values of Tcf7_Tcf7l2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_112368557 | 0.62 |
ENSRNOT00000075017
|
AABR07009787.1
|
|
| chr17_+_48436720 | 0.62 |
ENSRNOT00000074309
|
AABR07027902.1
|
|
| chr11_+_67555658 | 0.60 |
ENSRNOT00000039075
|
Csta
|
cystatin A (stefin A) |
| chr3_+_151691515 | 0.60 |
ENSRNOT00000052236
|
AC118414.1
|
|
| chr1_+_168668707 | 0.60 |
ENSRNOT00000015108
|
AC107531.3
|
|
| chr6_+_127941526 | 0.56 |
ENSRNOT00000033897
|
LOC500712
|
Ab1-233 |
| chr10_-_109840047 | 0.54 |
ENSRNOT00000054947
|
Notum
|
NOTUM, palmitoleoyl-protein carboxylesterase |
| chr3_-_76012929 | 0.50 |
ENSRNOT00000085199
|
AC111632.2
|
|
| chr15_-_41338284 | 0.45 |
ENSRNOT00000077225
|
Tnfrsf19
|
TNF receptor superfamily member 19 |
| chr17_+_43868191 | 0.44 |
ENSRNOT00000059403
|
Btn2a2
|
butyrophilin, subfamily 2, member A2 |
| chr10_-_88152064 | 0.40 |
ENSRNOT00000019477
|
Krt16
|
keratin 16 |
| chr9_+_12346117 | 0.37 |
ENSRNOT00000074599
|
AABR07066677.1
|
|
| chr2_+_71753816 | 0.36 |
ENSRNOT00000025809
|
AABR07008724.1
|
|
| chr9_+_12420368 | 0.36 |
ENSRNOT00000071620
|
AABR07066693.1
|
|
| chr17_+_22619891 | 0.35 |
ENSRNOT00000060403
|
Adtrp
|
androgen-dependent TFPI-regulating protein |
| chr6_+_106039991 | 0.34 |
ENSRNOT00000088917
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr1_-_16989482 | 0.34 |
ENSRNOT00000072759
|
AABR07000534.1
|
|
| chr2_-_170460754 | 0.34 |
ENSRNOT00000013009
|
Slitrk3
|
SLIT and NTRK-like family, member 3 |
| chr2_+_144861455 | 0.32 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr1_+_252409268 | 0.31 |
ENSRNOT00000026219
|
Lipm
|
lipase, family member M |
| chr19_-_21970103 | 0.28 |
ENSRNOT00000074210
|
AABR07043115.1
|
|
| chr7_-_15821927 | 0.27 |
ENSRNOT00000050658
|
LOC691422
|
similar to zinc finger protein 101 |
| chr4_+_22859622 | 0.27 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chr2_+_179952227 | 0.27 |
ENSRNOT00000015081
|
Pdgfc
|
platelet derived growth factor C |
| chr14_+_11988671 | 0.27 |
ENSRNOT00000003207
|
Rasgef1b
|
RasGEF domain family, member 1B |
| chr17_-_86873003 | 0.26 |
ENSRNOT00000061526
|
6720489N17Rik
|
RIKEN cDNA 6720489N17 gene |
| chr13_-_26768708 | 0.25 |
ENSRNOT00000092763
|
Bcl2
|
BCL2, apoptosis regulator |
| chr7_+_20462081 | 0.25 |
ENSRNOT00000088383
|
AABR07056131.1
|
|
| chr7_-_143538579 | 0.25 |
ENSRNOT00000081518
|
Krt79
|
keratin 79 |
| chr8_+_49441106 | 0.25 |
ENSRNOT00000030152
|
Scn4b
|
sodium voltage-gated channel beta subunit 4 |
| chr1_-_82004538 | 0.24 |
ENSRNOT00000087572
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr5_+_74766636 | 0.24 |
ENSRNOT00000030913
|
Rn50_5_0791.1
|
|
| chr3_-_2719135 | 0.24 |
ENSRNOT00000080257
|
Lcn12
|
lipocalin 12 |
| chr3_-_2719513 | 0.24 |
ENSRNOT00000020997
|
Lcn12
|
lipocalin 12 |
| chr5_+_48456757 | 0.23 |
ENSRNOT00000091482
|
Srsf12
|
serine and arginine rich splicing factor 12 |
| chr13_-_82005741 | 0.23 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr1_-_170186462 | 0.22 |
ENSRNOT00000035798
|
Olr202
|
olfactory receptor 202 |
| chr16_+_3851270 | 0.22 |
ENSRNOT00000014964
|
Plac9
|
placenta-specific 9 |
| chr13_-_82006005 | 0.22 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr6_+_108123816 | 0.22 |
ENSRNOT00000089242
ENSRNOT00000063900 |
Bbof1
|
basal body orientation factor 1 |
| chr14_-_92495894 | 0.21 |
ENSRNOT00000064483
|
Cobl
|
cordon-bleu WH2 repeat protein |
| chr4_+_102426224 | 0.21 |
ENSRNOT00000073768
|
LOC103694107
|
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3 pseudogene |
| chr1_+_128614138 | 0.21 |
ENSRNOT00000076227
ENSRNOT00000078707 |
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr1_+_61522298 | 0.21 |
ENSRNOT00000029111
|
Zfp51
|
zinc finger protein 51 |
| chr3_+_21764377 | 0.20 |
ENSRNOT00000063938
|
Gpr21
|
G protein-coupled receptor 21 |
| chr3_+_72329967 | 0.20 |
ENSRNOT00000090256
|
Slc43a3
|
solute carrier family 43, member 3 |
| chr10_-_37645802 | 0.20 |
ENSRNOT00000008022
|
Tcf7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr3_+_149741312 | 0.20 |
ENSRNOT00000070854
|
LOC100911637
|
high mobility group protein B1-like |
| chr10_-_40994872 | 0.20 |
ENSRNOT00000058640
|
RGD1563668
|
similar to High mobility group protein 1 (HMG-1) |
| chr3_-_153321352 | 0.19 |
ENSRNOT00000064824
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chr9_+_99795678 | 0.19 |
ENSRNOT00000056601
|
Olr1353
|
olfactory receptor 1353 |
| chr4_-_169999873 | 0.18 |
ENSRNOT00000011697
|
Grin2b
|
glutamate ionotropic receptor NMDA type subunit 2B |
| chr12_-_37682964 | 0.17 |
ENSRNOT00000001421
|
RGD1563482
|
similar to hypothetical protein FLJ38663 |
| chr8_+_44061894 | 0.17 |
ENSRNOT00000084416
|
Zfp202
|
zinc finger protein 202 |
| chr10_+_38104558 | 0.17 |
ENSRNOT00000067809
|
Fstl4
|
follistatin-like 4 |
| chr2_-_40386669 | 0.17 |
ENSRNOT00000014074
|
Elovl7
|
ELOVL fatty acid elongase 7 |
| chr8_+_64531594 | 0.17 |
ENSRNOT00000036798
|
Gramd2
|
GRAM domain containing 2 |
| chr7_+_141642777 | 0.17 |
ENSRNOT00000079811
|
AABR07058884.2
|
|
| chr3_-_25212049 | 0.16 |
ENSRNOT00000040023
|
Lrp1b
|
LDL receptor related protein 1B |
| chr1_-_219422268 | 0.16 |
ENSRNOT00000025092
|
Carns1
|
carnosine synthase 1 |
| chrX_-_25628272 | 0.16 |
ENSRNOT00000086414
|
Mid1
|
midline 1 |
| chr4_+_92431710 | 0.16 |
ENSRNOT00000049438
|
LOC108350839
|
high mobility group protein B1-like |
| chr1_-_248377093 | 0.16 |
ENSRNOT00000067612
|
Gldc
|
glycine decarboxylase |
| chr17_-_88095729 | 0.15 |
ENSRNOT00000025140
|
Enkur
|
enkurin, TRPC channel interacting protein |
| chr20_-_27247309 | 0.15 |
ENSRNOT00000037409
|
LOC100910755
|
RUN and FYVE domain-containing protein 2-like |
| chr13_-_107886476 | 0.15 |
ENSRNOT00000077282
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr1_+_81474553 | 0.14 |
ENSRNOT00000083493
|
Phldb3
|
pleckstrin homology-like domain, family B, member 3 |
| chr6_-_61405195 | 0.14 |
ENSRNOT00000008655
|
Lrrn3
|
leucine rich repeat neuronal 3 |
| chr3_-_44177689 | 0.14 |
ENSRNOT00000006387
|
Cytip
|
cytohesin 1 interacting protein |
| chr1_+_255710658 | 0.14 |
ENSRNOT00000031545
|
AC094246.1
|
|
| chr1_+_61786900 | 0.14 |
ENSRNOT00000090287
|
AABR07001905.1
|
|
| chr6_-_76652757 | 0.14 |
ENSRNOT00000042177
|
LOC100359668
|
ferritin light chain 2 |
| chr7_-_116201756 | 0.14 |
ENSRNOT00000091401
|
Cyp11b2
|
cytochrome P450, family 11, subfamily b, polypeptide 2 |
| chr9_+_18001105 | 0.14 |
ENSRNOT00000071814
|
AABR07066826.2
|
|
| chr4_-_161278293 | 0.13 |
ENSRNOT00000050332
|
AABR07062108.1
|
|
| chr15_-_64191115 | 0.13 |
ENSRNOT00000013868
|
LOC100361180
|
40S ribosomal protein S17-like |
| chr8_-_53758774 | 0.13 |
ENSRNOT00000068040
|
Ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr3_+_149790856 | 0.13 |
ENSRNOT00000075275
|
LOC100911637
|
high mobility group protein B1-like |
| chr13_+_83996080 | 0.13 |
ENSRNOT00000004403
ENSRNOT00000070958 |
Cd247
|
Cd247 molecule |
| chr9_-_9585865 | 0.13 |
ENSRNOT00000073926
|
Adgre1
|
adhesion G protein-coupled receptor E1 |
| chr12_-_51965779 | 0.13 |
ENSRNOT00000056733
|
LOC100362927
|
replication protein A3-like |
| chr8_+_71591360 | 0.13 |
ENSRNOT00000090096
|
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr4_+_150199627 | 0.13 |
ENSRNOT00000052259
|
AABR07061871.1
|
|
| chr8_-_47404010 | 0.13 |
ENSRNOT00000038647
|
Tmem136
|
transmembrane protein 136 |
| chr3_+_152226070 | 0.13 |
ENSRNOT00000073201
|
AABR07054397.1
|
|
| chr7_-_70661891 | 0.13 |
ENSRNOT00000010240
|
Inhbc
|
inhibin beta C subunit |
| chr7_-_140417530 | 0.13 |
ENSRNOT00000077884
|
Fkbp11
|
FK506 binding protein 11 |
| chr5_+_148066062 | 0.13 |
ENSRNOT00000084118
|
AABR07050006.1
|
|
| chr7_-_140090922 | 0.12 |
ENSRNOT00000014456
|
Lalba
|
lactalbumin, alpha |
| chr2_+_189993262 | 0.12 |
ENSRNOT00000016034
|
S100a3
|
S100 calcium binding protein A3 |
| chr6_-_122572925 | 0.12 |
ENSRNOT00000090869
|
AC123293.2
|
|
| chr6_-_111329882 | 0.12 |
ENSRNOT00000074480
|
Ism2
|
isthmin 2 |
| chr8_+_103774358 | 0.12 |
ENSRNOT00000014481
|
Xrn1
|
5'-3' exoribonuclease 1 |
| chr14_-_114047527 | 0.12 |
ENSRNOT00000083199
|
AABR07016779.1
|
|
| chr3_+_176494768 | 0.12 |
ENSRNOT00000040270
|
Col20a1
|
collagen type XX alpha 1 chain |
| chr4_+_108301129 | 0.12 |
ENSRNOT00000007993
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr1_-_52894832 | 0.11 |
ENSRNOT00000016471
|
T
|
T brachyury transcription factor |
| chr9_-_41996077 | 0.11 |
ENSRNOT00000087372
|
AABR07067387.1
|
|
| chr10_-_98294522 | 0.11 |
ENSRNOT00000005489
|
Abca8
|
ATP binding cassette subfamily A member 8 |
| chr12_-_9988618 | 0.11 |
ENSRNOT00000087449
|
Rasl11a
|
RAS-like family 11 member A |
| chr3_-_158328881 | 0.11 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr2_-_208633945 | 0.11 |
ENSRNOT00000049155
|
Pifo
|
primary cilia formation |
| chr3_-_146470293 | 0.11 |
ENSRNOT00000009627
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr18_+_87637608 | 0.11 |
ENSRNOT00000070851
|
LOC102550729
|
zinc finger protein 120-like |
| chr1_-_120893997 | 0.11 |
ENSRNOT00000081028
|
AABR07003933.1
|
|
| chr1_-_263431290 | 0.11 |
ENSRNOT00000022633
|
Slc25a28
|
solute carrier family 25 member 28 |
| chr5_+_74727494 | 0.11 |
ENSRNOT00000076683
|
Rn50_5_0791.1
|
|
| chrX_-_115375492 | 0.11 |
ENSRNOT00000007194
|
AABR07040944.1
|
|
| chr10_-_74119009 | 0.11 |
ENSRNOT00000085712
ENSRNOT00000006926 |
Dhx40
|
DEAH-box helicase 40 |
| chr15_-_82482009 | 0.11 |
ENSRNOT00000011926
|
Dach1
|
dachshund family transcription factor 1 |
| chr7_-_139835876 | 0.11 |
ENSRNOT00000014258
|
Olr1877
|
olfactory receptor 1877 |
| chr8_-_87315955 | 0.10 |
ENSRNOT00000081437
|
Filip1
|
filamin A interacting protein 1 |
| chr2_-_232373409 | 0.10 |
ENSRNOT00000041479
|
AABR07013302.1
|
|
| chr17_-_17947777 | 0.10 |
ENSRNOT00000036876
|
Rnf144b
|
ring finger protein 144B |
| chr14_-_2610929 | 0.10 |
ENSRNOT00000036442
|
Ccdc18
|
coiled-coil domain containing 18 |
| chr15_+_28439317 | 0.10 |
ENSRNOT00000060519
|
LOC690384
|
similar to ribosomal protein L31 |
| chr12_-_11808977 | 0.10 |
ENSRNOT00000071795
|
AABR07035375.1
|
|
| chrX_-_152642531 | 0.10 |
ENSRNOT00000085037
|
Gabra3
|
gamma-aminobutyric acid type A receptor alpha3 subunit |
| chr4_-_28310178 | 0.10 |
ENSRNOT00000084021
|
RGD1563091
|
similar to OEF2 |
| chr2_-_98610368 | 0.10 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr1_+_248210041 | 0.10 |
ENSRNOT00000047024
|
AABR07006688.2
|
|
| chr6_+_106084815 | 0.10 |
ENSRNOT00000058195
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr14_-_62646110 | 0.10 |
ENSRNOT00000002992
|
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr11_+_67101714 | 0.10 |
ENSRNOT00000078936
|
Cd86
|
CD86 molecule |
| chr8_+_94693290 | 0.10 |
ENSRNOT00000013908
|
Cyb5r4
|
cytochrome b5 reductase 4 |
| chr8_+_5606592 | 0.10 |
ENSRNOT00000011727
|
Mmp12
|
matrix metallopeptidase 12 |
| chr7_+_63352046 | 0.09 |
ENSRNOT00000046453
|
RGD1561871
|
similar to ribosomal protein S10 |
| chr9_+_60039297 | 0.09 |
ENSRNOT00000016262
|
Slc39a10
|
solute carrier family 39 member 10 |
| chr12_-_37688743 | 0.09 |
ENSRNOT00000030251
|
RGD1563482
|
similar to hypothetical protein FLJ38663 |
| chr17_+_88095830 | 0.09 |
ENSRNOT00000045984
|
Thnsl1
|
threonine synthase-like 1 |
| chr9_-_73799427 | 0.09 |
ENSRNOT00000067589
|
Kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr8_+_22648323 | 0.09 |
ENSRNOT00000013165
|
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr13_+_44475970 | 0.09 |
ENSRNOT00000024602
ENSRNOT00000091645 |
Ccnt2
|
cyclin T2 |
| chr8_+_58431407 | 0.09 |
ENSRNOT00000011974
|
Sln
|
sarcolipin |
| chr15_+_48674380 | 0.09 |
ENSRNOT00000018762
|
Fbxo16
|
F-box protein 16 |
| chr12_-_52596076 | 0.09 |
ENSRNOT00000056639
|
Chfr
|
checkpoint with forkhead and ring finger domains |
| chr11_+_20471764 | 0.09 |
ENSRNOT00000078127
|
Ncam2
|
neural cell adhesion molecule 2 |
| chr14_-_18862407 | 0.09 |
ENSRNOT00000003823
|
Cxcl6
|
C-X-C motif chemokine ligand 6 |
| chr5_-_6083083 | 0.09 |
ENSRNOT00000072411
|
Sulf1
|
sulfatase 1 |
| chr10_-_25910298 | 0.08 |
ENSRNOT00000065633
ENSRNOT00000079646 |
Ccng1
|
cyclin G1 |
| chr1_+_199163086 | 0.08 |
ENSRNOT00000025646
|
Ctf1
|
cardiotrophin 1 |
| chr12_+_19231092 | 0.08 |
ENSRNOT00000045379
|
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr19_+_56803382 | 0.08 |
ENSRNOT00000036042
|
Urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr18_+_46148849 | 0.08 |
ENSRNOT00000026724
|
Prr16
|
proline rich 16 |
| chr8_+_56451240 | 0.08 |
ENSRNOT00000085278
|
LOC100359687
|
mitochondrial ribosomal protein L1-like |
| chr15_-_34612432 | 0.08 |
ENSRNOT00000090206
|
Mcpt1l1
|
mast cell protease 1-like 1 |
| chr2_-_125323454 | 0.08 |
ENSRNOT00000045837
|
Ankrd50
|
ankyrin repeat domain 50 |
| chr13_-_90641772 | 0.08 |
ENSRNOT00000064601
|
Atp1a4
|
ATPase Na+/K+ transporting subunit alpha 4 |
| chr12_-_13374500 | 0.08 |
ENSRNOT00000038577
|
Zfp12
|
zinc finger protein 12 |
| chr13_+_70157522 | 0.08 |
ENSRNOT00000036906
|
Apobec4
|
apolipoprotein B mRNA editing enzyme catalytic polypeptide like 4 |
| chr1_-_62316450 | 0.08 |
ENSRNOT00000079171
|
AABR07001926.3
|
|
| chrX_-_21710341 | 0.08 |
ENSRNOT00000091261
|
Ribc1
|
RIB43A domain with coiled-coils 1 |
| chr3_-_46726946 | 0.08 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr1_+_224957517 | 0.08 |
ENSRNOT00000026033
|
Nxf1
|
nuclear RNA export factor 1 |
| chr1_+_80973739 | 0.08 |
ENSRNOT00000026088
ENSRNOT00000081983 |
AC118165.1
|
|
| chr10_+_89285855 | 0.08 |
ENSRNOT00000028033
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chrX_+_135470915 | 0.08 |
ENSRNOT00000009637
|
Slc25a14
|
solute carrier family 25 member 14 |
| chr2_+_207351051 | 0.08 |
ENSRNOT00000084302
|
St7l
|
suppression of tumorigenicity 7-like |
| chr5_-_77294735 | 0.08 |
ENSRNOT00000018209
ENSRNOT00000056900 |
Zfp37
|
zinc finger protein 37 |
| chr1_-_141579871 | 0.08 |
ENSRNOT00000020002
|
Anpep
|
alanyl aminopeptidase, membrane |
| chr5_-_147907733 | 0.08 |
ENSRNOT00000032558
|
Tmem39b
|
transmembrane protein 39b |
| chr17_+_45078556 | 0.08 |
ENSRNOT00000088280
|
Zfp192
|
zinc finger protein 192 |
| chr7_+_35070284 | 0.08 |
ENSRNOT00000009578
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr9_-_17065600 | 0.07 |
ENSRNOT00000025638
|
Dlk2
|
delta like non-canonical Notch ligand 2 |
| chr1_+_187149453 | 0.07 |
ENSRNOT00000082738
|
Xylt1
|
xylosyltransferase 1 |
| chr5_+_126254142 | 0.07 |
ENSRNOT00000057471
|
Pars2
|
prolyl-tRNA synthetase 2, mitochondrial (putative) |
| chr11_+_66878658 | 0.07 |
ENSRNOT00000003208
|
Eaf2
|
ELL associated factor 2 |
| chr10_-_74376270 | 0.07 |
ENSRNOT00000079311
|
Gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr1_-_31936537 | 0.07 |
ENSRNOT00000060971
|
Zdhhc11
|
zinc finger, DHHC-type containing 11 |
| chr4_+_179481263 | 0.07 |
ENSRNOT00000021284
|
Etfrf1
|
electron transfer flavoprotein regulatory factor 1 |
| chr4_-_159697207 | 0.07 |
ENSRNOT00000086440
|
Ccnd2
|
cyclin D2 |
| chr10_+_43446526 | 0.07 |
ENSRNOT00000036959
|
Larp1
|
La ribonucleoprotein domain family, member 1 |
| chr13_+_63526486 | 0.07 |
ENSRNOT00000003788
|
Brinp3
|
BMP/retinoic acid inducible neural specific 3 |
| chr1_+_185863043 | 0.07 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr2_+_243422811 | 0.07 |
ENSRNOT00000014694
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr1_-_88162583 | 0.07 |
ENSRNOT00000087411
|
Catsperg
|
cation channel sperm associated auxiliary subunit gamma |
| chr6_-_25211494 | 0.07 |
ENSRNOT00000009634
|
Xdh
|
xanthine dehydrogenase |
| chr6_+_73553210 | 0.07 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr1_+_88457594 | 0.07 |
ENSRNOT00000049877
|
Zfp260
|
zinc finger protein 260 |
| chr10_+_74413989 | 0.07 |
ENSRNOT00000036098
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr2_-_91497091 | 0.07 |
ENSRNOT00000015185
|
Pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr9_+_92681078 | 0.07 |
ENSRNOT00000034152
|
Sp100
|
SP100 nuclear antigen |
| chr1_-_218100272 | 0.07 |
ENSRNOT00000028411
ENSRNOT00000088588 |
Ccnd1
|
cyclin D1 |
| chr11_+_88732381 | 0.07 |
ENSRNOT00000078367
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr1_+_88922433 | 0.07 |
ENSRNOT00000045212
|
Nphs1
|
NPHS1 nephrin |
| chr3_+_8451292 | 0.07 |
ENSRNOT00000020407
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr3_-_166993940 | 0.07 |
ENSRNOT00000034669
|
Zfp217
|
zinc finger protein 217 |
| chr15_-_20822740 | 0.07 |
ENSRNOT00000012957
|
Bmp4
|
bone morphogenetic protein 4 |
| chr1_-_62558033 | 0.07 |
ENSRNOT00000015520
|
Zfp40
|
zinc finger protein 40 |
| chr1_+_202432366 | 0.06 |
ENSRNOT00000027681
|
Plpp4
|
phospholipid phosphatase 4 |
| chr17_+_9736577 | 0.06 |
ENSRNOT00000066586
|
F12
|
coagulation factor XII |
| chr4_+_7144669 | 0.06 |
ENSRNOT00000077682
ENSRNOT00000014998 |
Iqca1l
|
IQ motif containing with AAA domain 1 like |
| chr8_-_87419564 | 0.06 |
ENSRNOT00000015365
|
Filip1
|
filamin A interacting protein 1 |
| chrX_-_21710678 | 0.06 |
ENSRNOT00000004184
|
Ribc1
|
RIB43A domain with coiled-coils 1 |
| chr4_-_161091010 | 0.06 |
ENSRNOT00000043708
|
Senp17
|
Sumo1/sentrin/SMT3 specific peptidase 17 |
| chr8_+_76400341 | 0.06 |
ENSRNOT00000087382
ENSRNOT00000081072 |
Bnip2
|
BCL2/adenovirus E1B interacting protein 2 |
| chr8_+_107882219 | 0.06 |
ENSRNOT00000019902
|
Dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr20_-_4390436 | 0.06 |
ENSRNOT00000000497
ENSRNOT00000077655 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr11_-_61530567 | 0.06 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr18_-_786674 | 0.06 |
ENSRNOT00000021955
|
Cetn1
|
centrin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf7_Tcf7l2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 0.3 | GO:0046671 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 0.2 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.1 | 0.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.5 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.2 | GO:0048669 | somite specification(GO:0001757) collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.2 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 0.0 | 0.2 | GO:1902951 | NMDA selective glutamate receptor signaling pathway(GO:0098989) negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.0 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.1 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.2 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.2 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.1 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:0072642 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.4 | GO:0051546 | keratinization(GO:0031424) keratinocyte migration(GO:0051546) |
| 0.0 | 0.1 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.0 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 0.0 | GO:1990478 | response to ultrasound(GO:1990478) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.3 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:1900747 | xanthine catabolic process(GO:0009115) negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0086016 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.0 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.0 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.0 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.0 | GO:0009080 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.0 | 0.0 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.0 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.0 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.2 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.1 | GO:0032551 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.0 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.0 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.0 | 0.0 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |