Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Tcf4_Mesp1
Z-value: 0.35
Transcription factors associated with Tcf4_Mesp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
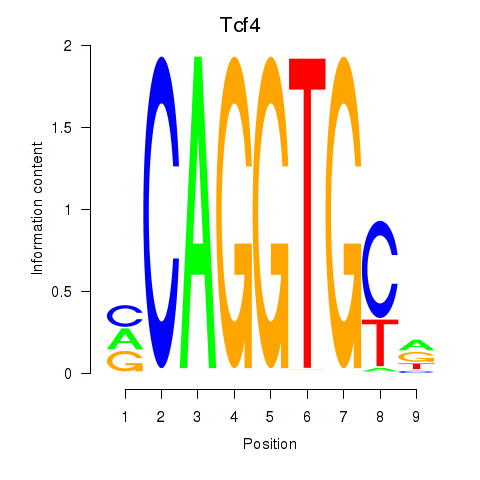
Tcf4
|
ENSRNOG00000012405 | transcription factor 4 |
|
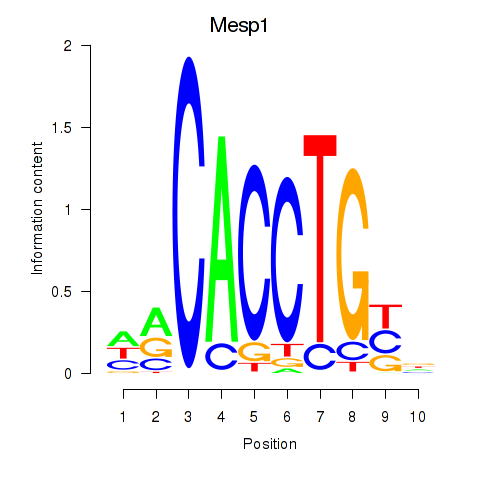
Mesp1
|
ENSRNOG00000014951 | mesoderm posterior bHLH transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf4 | rn6_v1_chr18_+_65285318_65285318 | -0.35 | 5.7e-01 | Click! |
| Mesp1 | rn6_v1_chr1_-_141533908_141533908 | -0.23 | 7.1e-01 | Click! |
Activity profile of Tcf4_Mesp1 motif
Sorted Z-values of Tcf4_Mesp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_2174131 | 0.79 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr6_+_33885495 | 0.43 |
ENSRNOT00000086633
|
Sdc1
|
syndecan 1 |
| chr10_+_59529785 | 0.33 |
ENSRNOT00000064840
ENSRNOT00000065181 |
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr7_-_130120579 | 0.28 |
ENSRNOT00000044376
|
Mapk12
|
mitogen-activated protein kinase 12 |
| chr5_-_21345805 | 0.28 |
ENSRNOT00000081296
ENSRNOT00000007802 |
Car8
|
carbonic anhydrase 8 |
| chr16_-_20807070 | 0.27 |
ENSRNOT00000072536
|
Comp
|
cartilage oligomeric matrix protein |
| chr5_+_154522119 | 0.25 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr3_-_9037942 | 0.25 |
ENSRNOT00000036770
|
Ier5l
|
immediate early response 5-like |
| chr19_-_46167950 | 0.24 |
ENSRNOT00000071024
|
AABR07043892.1
|
|
| chr3_-_123119460 | 0.24 |
ENSRNOT00000028833
|
Avp
|
arginine vasopressin |
| chr9_+_16565225 | 0.21 |
ENSRNOT00000022307
|
Gnmt
|
glycine N-methyltransferase |
| chr9_-_9985630 | 0.21 |
ENSRNOT00000071780
|
Crb3
|
crumbs 3, cell polarity complex component |
| chr1_+_221673590 | 0.21 |
ENSRNOT00000038016
|
Cdc42bpg
|
CDC42 binding protein kinase gamma |
| chr7_-_107634287 | 0.19 |
ENSRNOT00000093672
ENSRNOT00000087116 |
Sla
|
src-like adaptor |
| chr18_-_3662654 | 0.18 |
ENSRNOT00000092854
ENSRNOT00000016167 |
Npc1
|
NPC intracellular cholesterol transporter 1 |
| chr19_+_41482728 | 0.18 |
ENSRNOT00000022943
|
Calb2
|
calbindin 2 |
| chr20_+_40618128 | 0.17 |
ENSRNOT00000001075
ENSRNOT00000057364 |
Pkib
|
cAMP-dependent protein kinase inhibitor beta |
| chr9_+_84203072 | 0.16 |
ENSRNOT00000018882
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chr5_+_117052260 | 0.15 |
ENSRNOT00000010211
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr16_-_19918644 | 0.15 |
ENSRNOT00000083345
ENSRNOT00000023926 |
Plvap
|
plasmalemma vesicle associated protein |
| chr10_-_88551056 | 0.15 |
ENSRNOT00000032737
|
Zfp385c
|
zinc finger protein 385C |
| chr10_+_86399827 | 0.15 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr6_-_99783047 | 0.15 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr16_-_20888907 | 0.15 |
ENSRNOT00000027428
|
Homer3
|
homer scaffolding protein 3 |
| chr20_+_5067330 | 0.15 |
ENSRNOT00000037482
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr9_+_16314173 | 0.15 |
ENSRNOT00000072812
|
LOC100911564
|
glycine N-methyltransferase-like |
| chr6_-_11494459 | 0.14 |
ENSRNOT00000021570
|
Kcnk12
|
potassium two pore domain channel subfamily K member 12 |
| chr7_-_123445613 | 0.14 |
ENSRNOT00000070937
|
Shisa8
|
shisa family member 8 |
| chrX_-_111942749 | 0.14 |
ENSRNOT00000087583
|
AABR07040840.1
|
|
| chr12_-_48857724 | 0.13 |
ENSRNOT00000080783
|
Wscd2
|
WSC domain containing 2 |
| chr6_+_3012804 | 0.13 |
ENSRNOT00000061980
|
Arhgef33
|
Rho guanine nucleotide exchange factor 33 |
| chr14_-_82055290 | 0.13 |
ENSRNOT00000058062
|
RGD1560394
|
RGD1560394 |
| chrX_-_157286936 | 0.12 |
ENSRNOT00000078100
|
Atp2b3
|
ATPase plasma membrane Ca2+ transporting 3 |
| chr1_-_264975132 | 0.12 |
ENSRNOT00000021748
|
Lbx1
|
ladybird homeobox 1 |
| chr20_-_3440769 | 0.12 |
ENSRNOT00000084981
|
Ier3
|
immediate early response 3 |
| chr13_+_25778317 | 0.12 |
ENSRNOT00000021129
|
Tnfrsf11a
|
TNF receptor superfamily member 11A |
| chr17_-_84488480 | 0.12 |
ENSRNOT00000000158
ENSRNOT00000075983 |
Nebl
|
nebulette |
| chr19_-_645937 | 0.12 |
ENSRNOT00000016521
|
Car7
|
carbonic anhydrase 7 |
| chr7_+_11737293 | 0.12 |
ENSRNOT00000046059
|
Lingo3
|
leucine rich repeat and Ig domain containing 3 |
| chr4_-_160662974 | 0.12 |
ENSRNOT00000089199
|
Tspan9
|
tetraspanin 9 |
| chr8_+_57886168 | 0.11 |
ENSRNOT00000039336
|
Exph5
|
exophilin 5 |
| chr20_+_6351458 | 0.11 |
ENSRNOT00000091731
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr15_-_28406046 | 0.11 |
ENSRNOT00000015418
|
Zfp219
|
zinc finger protein 219 |
| chr12_+_47551935 | 0.11 |
ENSRNOT00000056932
|
RGD1560398
|
RGD1560398 |
| chr9_-_93735636 | 0.11 |
ENSRNOT00000025582
|
Nppc
|
natriuretic peptide C |
| chr12_-_16934706 | 0.11 |
ENSRNOT00000001720
|
Mafk
|
MAF bZIP transcription factor K |
| chrX_-_156155014 | 0.11 |
ENSRNOT00000088637
|
LOC102552182
|
L antigen family member 3-like |
| chr1_-_142197182 | 0.10 |
ENSRNOT00000015521
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr16_-_59366824 | 0.10 |
ENSRNOT00000015062
|
RGD1304810
|
similar to 6430573F11Rik protein |
| chr4_-_82160240 | 0.10 |
ENSRNOT00000038775
ENSRNOT00000058985 |
Hoxa4
|
homeo box A4 |
| chr19_-_43596801 | 0.10 |
ENSRNOT00000025625
|
Fa2h
|
fatty acid 2-hydroxylase |
| chr6_-_102196138 | 0.10 |
ENSRNOT00000014132
|
Tmem229b
|
transmembrane protein 229B |
| chr7_-_101140308 | 0.10 |
ENSRNOT00000006279
|
Fam84b
|
family with sequence similarity 84, member B |
| chr10_+_47281786 | 0.10 |
ENSRNOT00000089123
|
Kcnj12
|
potassium voltage-gated channel subfamily J member 12 |
| chr15_-_30147793 | 0.10 |
ENSRNOT00000060399
|
AABR07017693.1
|
|
| chr20_-_12352377 | 0.10 |
ENSRNOT00000044212
|
Slc19a1
|
solute carrier family 19 member 1 |
| chr8_+_73593310 | 0.10 |
ENSRNOT00000012048
|
C2cd4b
|
C2 calcium-dependent domain containing 4B |
| chr1_-_89539210 | 0.10 |
ENSRNOT00000077462
ENSRNOT00000028644 |
Hpn
|
hepsin |
| chr1_+_105284753 | 0.09 |
ENSRNOT00000041950
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr20_-_13706205 | 0.09 |
ENSRNOT00000038623
|
Derl3
|
derlin 3 |
| chr6_-_125027553 | 0.09 |
ENSRNOT00000005908
|
Ccdc88c
|
coiled-coil domain containing 88C |
| chr8_+_130749838 | 0.09 |
ENSRNOT00000079273
|
Snrk
|
SNF related kinase |
| chr14_+_16491573 | 0.09 |
ENSRNOT00000002995
|
Sowahb
|
sosondowah ankyrin repeat domain family member B |
| chr7_+_144647587 | 0.09 |
ENSRNOT00000022398
|
Hoxc4
|
homeo box C4 |
| chr8_-_108109801 | 0.09 |
ENSRNOT00000034277
|
Sox14
|
SRY box 14 |
| chr8_-_115167486 | 0.09 |
ENSRNOT00000033018
|
Gpr62
|
G protein-coupled receptor 62 |
| chr3_+_2877293 | 0.09 |
ENSRNOT00000061855
|
Lcn5
|
lipocalin 5 |
| chr15_+_37806836 | 0.09 |
ENSRNOT00000076285
|
Il17d
|
interleukin 17D |
| chr9_+_40972089 | 0.09 |
ENSRNOT00000067928
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr12_+_52637000 | 0.08 |
ENSRNOT00000063905
|
Gtpbp6
|
GTP binding protein 6 (putative) |
| chr6_+_28382962 | 0.08 |
ENSRNOT00000016976
|
Pomc
|
proopiomelanocortin |
| chr15_-_80713153 | 0.08 |
ENSRNOT00000063800
|
Klhl1
|
kelch-like family member 1 |
| chr5_+_151776004 | 0.08 |
ENSRNOT00000009683
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr5_-_152987211 | 0.08 |
ENSRNOT00000000163
|
Ldlrap1
|
low density lipoprotein receptor adaptor protein 1 |
| chr5_+_139963002 | 0.08 |
ENSRNOT00000048506
|
Col9a2
|
collagen type IX alpha 2 chain |
| chr1_+_87224677 | 0.08 |
ENSRNOT00000028070
|
Ppp1r14a
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr18_+_76770012 | 0.08 |
ENSRNOT00000089389
|
Pqlc1
|
PQ loop repeat containing 1 |
| chr14_+_87312203 | 0.08 |
ENSRNOT00000088032
|
Adcy1
|
adenylate cyclase 1 |
| chr1_-_124803363 | 0.08 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr13_-_44345735 | 0.08 |
ENSRNOT00000005006
|
Tmem163
|
transmembrane protein 163 |
| chr3_+_161121697 | 0.08 |
ENSRNOT00000036020
|
Wfdc10a
|
WAP four-disulfide core domain 10A |
| chr5_-_12541019 | 0.08 |
ENSRNOT00000091649
|
St18
|
suppression of tumorigenicity 18 |
| chr6_-_109004598 | 0.08 |
ENSRNOT00000007790
|
Pgf
|
placental growth factor |
| chr17_-_27112820 | 0.08 |
ENSRNOT00000018359
|
Bmp6
|
bone morphogenetic protein 6 |
| chr15_-_33527031 | 0.08 |
ENSRNOT00000019979
|
Homez
|
homeobox and leucine zipper encoding |
| chr7_+_140742418 | 0.08 |
ENSRNOT00000089211
|
Prph
|
peripherin |
| chr16_+_20672374 | 0.07 |
ENSRNOT00000063780
|
RGD1566239
|
similar to RIKEN cDNA 2810428I15 |
| chr1_-_165680176 | 0.07 |
ENSRNOT00000025245
ENSRNOT00000082697 |
Plekhb1
|
pleckstrin homology domain containing B1 |
| chr10_+_48240330 | 0.07 |
ENSRNOT00000057798
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr7_-_130350570 | 0.07 |
ENSRNOT00000055805
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr4_-_123494742 | 0.07 |
ENSRNOT00000073268
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr9_-_9985358 | 0.07 |
ENSRNOT00000080856
|
Crb3
|
crumbs 3, cell polarity complex component |
| chr10_-_88307132 | 0.07 |
ENSRNOT00000040845
|
Jup
|
junction plakoglobin |
| chr15_+_41069507 | 0.07 |
ENSRNOT00000018533
|
C1qtnf9
|
C1q and tumor necrosis factor related protein 9 |
| chr1_+_211423022 | 0.07 |
ENSRNOT00000029587
|
Dpysl4
|
dihydropyrimidinase-like 4 |
| chr1_+_217173199 | 0.07 |
ENSRNOT00000075078
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr6_-_125723944 | 0.07 |
ENSRNOT00000007004
|
Fbln5
|
fibulin 5 |
| chr9_-_46401911 | 0.07 |
ENSRNOT00000046557
|
Creg2
|
cellular repressor of E1A-stimulated genes 2 |
| chr3_+_171037957 | 0.07 |
ENSRNOT00000008764
|
Rbm38
|
RNA binding motif protein 38 |
| chr4_+_180123905 | 0.07 |
ENSRNOT00000083173
|
Rassf8
|
Ras association domain family member 8 |
| chr14_+_83724933 | 0.07 |
ENSRNOT00000029848
|
Pla2g3
|
phospholipase A2, group III |
| chr1_+_100199057 | 0.07 |
ENSRNOT00000025831
|
Klk1
|
kallikrein 1 |
| chr1_-_89399039 | 0.07 |
ENSRNOT00000028585
ENSRNOT00000044678 |
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr10_-_105116916 | 0.07 |
ENSRNOT00000012373
|
Evpl
|
envoplakin |
| chr1_-_89068348 | 0.07 |
ENSRNOT00000033621
|
Upk1a
|
uroplakin 1A |
| chr5_+_169475270 | 0.07 |
ENSRNOT00000014625
|
Icmt
|
isoprenylcysteine carboxyl methyltransferase |
| chr15_+_30413488 | 0.06 |
ENSRNOT00000071781
|
AABR07017733.1
|
|
| chr1_+_214328071 | 0.06 |
ENSRNOT00000024725
|
Eps8l2
|
EPS8-like 2 |
| chr8_-_23390773 | 0.06 |
ENSRNOT00000019601
ENSRNOT00000071846 |
Anln
|
anillin, actin binding protein |
| chr7_-_12221330 | 0.06 |
ENSRNOT00000042204
|
Plk5
|
polo-like kinase 5 |
| chr1_+_152072665 | 0.06 |
ENSRNOT00000022538
|
Rab38
|
RAB38, member RAS oncogene family |
| chr10_-_82887497 | 0.06 |
ENSRNOT00000005644
|
Itga3
|
integrin subunit alpha 3 |
| chr14_-_85191557 | 0.06 |
ENSRNOT00000011604
|
Nefh
|
neurofilament heavy |
| chr12_-_44279002 | 0.06 |
ENSRNOT00000064900
|
Fbxo21
|
F-box protein 21 |
| chr12_-_11785493 | 0.06 |
ENSRNOT00000039687
|
Zfp498
|
zinc finger protein 498 |
| chr8_+_5967463 | 0.06 |
ENSRNOT00000086251
|
Tmem123
|
transmembrane protein 123 |
| chr10_+_66099531 | 0.06 |
ENSRNOT00000056192
|
Lyrm9
|
LYR motif containing 9 |
| chr17_-_72046381 | 0.06 |
ENSRNOT00000073919
|
AABR07028488.1
|
|
| chr12_+_52670898 | 0.06 |
ENSRNOT00000056632
|
LOC100912590
|
putative GTP-binding protein 6-like |
| chr19_-_25288335 | 0.06 |
ENSRNOT00000050214
|
Nanos3
|
nanos C2HC-type zinc finger 3 |
| chr1_+_24900858 | 0.06 |
ENSRNOT00000075616
|
LOC501467
|
similar to spermatogenesis associated glutamate (E)-rich protein 4d |
| chr5_-_137372524 | 0.06 |
ENSRNOT00000009061
|
Tmem125
|
transmembrane protein 125 |
| chr20_+_13778178 | 0.06 |
ENSRNOT00000058314
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr5_-_134978125 | 0.06 |
ENSRNOT00000018004
|
Rad54l
|
RAD54 like (S. cerevisiae) |
| chr6_-_125723732 | 0.06 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr19_+_16417817 | 0.06 |
ENSRNOT00000015272
|
Irx5
|
iroquois homeobox 5 |
| chr3_-_11417546 | 0.06 |
ENSRNOT00000018776
|
Lcn2
|
lipocalin 2 |
| chr14_+_80403001 | 0.06 |
ENSRNOT00000012109
|
Cpz
|
carboxypeptidase Z |
| chr5_-_12563429 | 0.06 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr1_+_46779860 | 0.06 |
ENSRNOT00000075268
|
Snx9
|
sorting nexin 9 |
| chr5_-_160620334 | 0.06 |
ENSRNOT00000019171
|
Tmem51
|
transmembrane protein 51 |
| chr10_-_108691367 | 0.06 |
ENSRNOT00000005067
|
Nptx1
|
neuronal pentraxin 1 |
| chr7_-_11003785 | 0.06 |
ENSRNOT00000061279
|
Tle2
|
transducin-like enhancer of split 2 |
| chr10_-_63952726 | 0.06 |
ENSRNOT00000090461
|
Doc2b
|
double C2 domain beta |
| chr19_+_25123724 | 0.06 |
ENSRNOT00000007407
|
LOC686013
|
hypothetical protein LOC686013 |
| chrX_+_71272042 | 0.06 |
ENSRNOT00000076034
ENSRNOT00000076816 |
Gjb1
|
gap junction protein, beta 1 |
| chr3_+_164424515 | 0.06 |
ENSRNOT00000083876
|
Cebpb
|
CCAAT/enhancer binding protein beta |
| chr8_-_130429132 | 0.06 |
ENSRNOT00000026261
|
Hhatl
|
hedgehog acyltransferase-like |
| chr9_-_98551410 | 0.06 |
ENSRNOT00000066346
ENSRNOT00000086678 |
Hes6
|
hes family bHLH transcription factor 6 |
| chr20_-_12351813 | 0.06 |
ENSRNOT00000065236
|
Slc19a1
|
solute carrier family 19 member 1 |
| chr10_+_40247436 | 0.06 |
ENSRNOT00000079830
|
Gpx3
|
glutathione peroxidase 3 |
| chr10_-_85938085 | 0.06 |
ENSRNOT00000043561
|
Plxdc1
|
plexin domain containing 1 |
| chr6_-_28994519 | 0.06 |
ENSRNOT00000006668
|
Ubxn2a
|
UBX domain protein 2A |
| chr1_+_79727597 | 0.05 |
ENSRNOT00000034790
|
AC093995.1
|
|
| chr5_-_127273656 | 0.05 |
ENSRNOT00000057341
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr8_+_116771982 | 0.05 |
ENSRNOT00000083240
|
AC128059.3
|
|
| chr19_+_27835890 | 0.05 |
ENSRNOT00000042150
|
LOC100909409
|
disks large homolog 5-like |
| chr15_+_37790141 | 0.05 |
ENSRNOT00000076392
ENSRNOT00000091953 |
Il17d
|
interleukin 17D |
| chr8_+_50559126 | 0.05 |
ENSRNOT00000024918
|
Apoa5
|
apolipoprotein A5 |
| chr20_+_4823662 | 0.05 |
ENSRNOT00000090028
|
Nfkbil1
|
NFKB inhibitor like 1 |
| chr10_+_36099263 | 0.05 |
ENSRNOT00000083568
|
Adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr10_-_38838272 | 0.05 |
ENSRNOT00000089495
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr11_+_71872830 | 0.05 |
ENSRNOT00000050364
|
Nrros
|
negative regulator of reactive oxygen species |
| chr10_-_91291774 | 0.05 |
ENSRNOT00000004356
|
LOC100361655
|
rCG33642-like |
| chrX_-_123662350 | 0.05 |
ENSRNOT00000092624
|
Sept6
|
septin 6 |
| chr9_+_10339075 | 0.05 |
ENSRNOT00000073402
|
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr4_+_87026530 | 0.05 |
ENSRNOT00000018425
|
Avl9
|
AVL9 cell migration associated |
| chr1_-_88558387 | 0.05 |
ENSRNOT00000074174
|
AABR07002896.1
|
|
| chr19_+_209926 | 0.05 |
ENSRNOT00000071393
|
LOC108348293
|
uncharacterized LOC108348293 |
| chr12_-_23841049 | 0.05 |
ENSRNOT00000031555
|
Hspb1
|
heat shock protein family B (small) member 1 |
| chr8_-_116349896 | 0.05 |
ENSRNOT00000087306
|
Lsmem2
|
leucine-rich single-pass membrane protein 2 |
| chr13_-_51183269 | 0.05 |
ENSRNOT00000039540
|
Ppfia4
|
PTPRF interacting protein alpha 4 |
| chr7_+_141370491 | 0.05 |
ENSRNOT00000087662
|
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 |
| chr20_-_3819200 | 0.05 |
ENSRNOT00000000542
ENSRNOT00000081486 |
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr7_-_11018160 | 0.05 |
ENSRNOT00000092061
|
Aes
|
amino-terminal enhancer of split |
| chr6_+_108285822 | 0.05 |
ENSRNOT00000015889
|
Vsx2
|
visual system homeobox 2 |
| chr5_+_137371825 | 0.05 |
ENSRNOT00000072816
|
Tmem125
|
transmembrane protein 125 |
| chr2_+_225827504 | 0.05 |
ENSRNOT00000018343
|
Gclm
|
glutamate cysteine ligase, modifier subunit |
| chr4_-_30556814 | 0.05 |
ENSRNOT00000012760
|
Pdk4
|
pyruvate dehydrogenase kinase 4 |
| chr1_-_277768368 | 0.05 |
ENSRNOT00000023113
|
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr1_+_221792221 | 0.05 |
ENSRNOT00000054828
|
Nrxn2
|
neurexin 2 |
| chr1_+_214440503 | 0.05 |
ENSRNOT00000025964
|
Cracr2b
|
calcium release activated channel regulator 2B |
| chr1_-_222118459 | 0.05 |
ENSRNOT00000067217
|
Ccdc88b
|
coiled-coil domain containing 88B |
| chr1_-_87221826 | 0.05 |
ENSRNOT00000046611
ENSRNOT00000028006 |
Spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr1_-_255815733 | 0.05 |
ENSRNOT00000047387
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr5_+_169452493 | 0.05 |
ENSRNOT00000014444
|
Gpr153
|
G protein-coupled receptor 153 |
| chr1_-_242765807 | 0.05 |
ENSRNOT00000020763
|
Pgm5
|
phosphoglucomutase 5 |
| chr1_-_216156409 | 0.05 |
ENSRNOT00000027688
|
Ascl2
|
achaete-scute family bHLH transcription factor 2 |
| chr7_-_98960736 | 0.05 |
ENSRNOT00000011984
ENSRNOT00000080155 |
Mtss1
|
MTSS1, I-BAR domain containing |
| chr6_+_110410141 | 0.05 |
ENSRNOT00000013867
|
Esrrb
|
estrogen-related receptor beta |
| chr4_-_31730386 | 0.05 |
ENSRNOT00000013817
|
Slc25a13
|
solute carrier family 25 member 13 |
| chr3_+_11593655 | 0.05 |
ENSRNOT00000074122
|
Pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr1_-_72366455 | 0.05 |
ENSRNOT00000031786
|
LOC100911196
|
zinc finger protein 865-like |
| chr12_+_41385241 | 0.05 |
ENSRNOT00000074974
|
Dtx1
|
deltex E3 ubiquitin ligase 1 |
| chr12_+_18074033 | 0.05 |
ENSRNOT00000001727
|
LOC103689975
|
integrator complex subunit 1-like |
| chr1_-_165891627 | 0.05 |
ENSRNOT00000038408
|
Relt
|
RELT tumor necrosis factor receptor |
| chr19_-_26053762 | 0.05 |
ENSRNOT00000004646
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr20_-_3386048 | 0.05 |
ENSRNOT00000089550
ENSRNOT00000065025 |
Dhx16
|
DEAH-box helicase 16 |
| chr18_+_38847632 | 0.05 |
ENSRNOT00000014916
|
LOC102554034
|
ELMO domain-containing protein 2-like |
| chr15_+_24153602 | 0.05 |
ENSRNOT00000014216
|
Lgals3
|
galectin 3 |
| chr4_+_156752082 | 0.05 |
ENSRNOT00000084588
ENSRNOT00000068407 |
Cd163
|
CD163 molecule |
| chr11_+_47200029 | 0.05 |
ENSRNOT00000079598
|
Nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr4_+_155313671 | 0.04 |
ENSRNOT00000020812
|
Mfap5
|
microfibrillar associated protein 5 |
| chr5_+_103479767 | 0.04 |
ENSRNOT00000008999
|
Sh3gl2
|
SH3 domain-containing GRB2-like 2 |
| chr8_-_22974321 | 0.04 |
ENSRNOT00000017369
|
Epor
|
erythropoietin receptor |
| chr4_-_183293999 | 0.04 |
ENSRNOT00000079695
|
Ipo8
|
importin 8 |
| chr12_-_40227297 | 0.04 |
ENSRNOT00000030600
|
Fam109a
|
family with sequence similarity 109, member A |
| chr5_+_28480023 | 0.04 |
ENSRNOT00000075067
ENSRNOT00000093754 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr4_+_7355574 | 0.04 |
ENSRNOT00000013800
|
Kcnh2
|
potassium voltage-gated channel subfamily H member 2 |
| chr17_+_15845931 | 0.04 |
ENSRNOT00000092083
|
Card19
|
caspase recruitment domain family, member 19 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf4_Mesp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.4 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.3 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.1 | 0.2 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.1 | GO:0032900 | negative regulation of neurotrophin production(GO:0032900) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.3 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.0 | 0.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:1990792 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.0 | 0.2 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:1903769 | negative regulation of cell proliferation in bone marrow(GO:1903769) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0035711 | granuloma formation(GO:0002432) T-helper 1 cell activation(GO:0035711) |
| 0.0 | 0.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.1 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.0 | GO:0071663 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.0 | 0.0 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.0 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:1904373 | response to kainic acid(GO:1904373) |
| 0.0 | 0.0 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.0 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.0 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.0 | 0.0 | GO:0031439 | positive regulation of mRNA cleavage(GO:0031439) positive regulation of endoribonuclease activity(GO:1902380) positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904722) |
| 0.0 | 0.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.0 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.0 | 0.1 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.0 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.0 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.0 | 0.0 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.0 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.0 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.0 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0032173 | septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.0 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.1 | 0.2 | GO:0098603 | selenol Se-methyltransferase activity(GO:0098603) |
| 0.1 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.2 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.0 | GO:0031249 | denatured protein binding(GO:0031249) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.0 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.0 | GO:0004956 | prostaglandin D receptor activity(GO:0004956) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |