Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
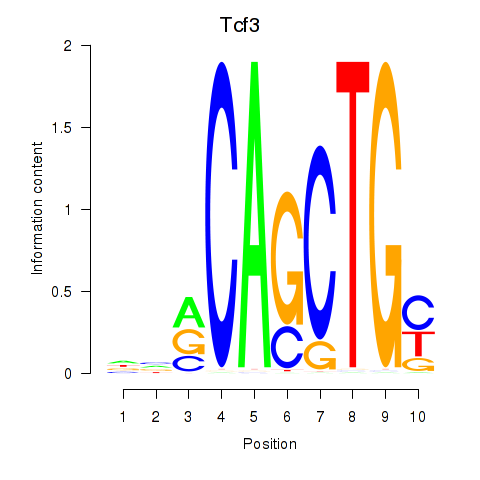
Results for Tcf3
Z-value: 0.34
Transcription factors associated with Tcf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf3
|
ENSRNOG00000051499 | transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf3 | rn6_v1_chr7_+_12146642_12146642 | 0.51 | 3.8e-01 | Click! |
Activity profile of Tcf3 motif
Sorted Z-values of Tcf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_9037942 | 0.25 |
ENSRNOT00000036770
|
Ier5l
|
immediate early response 5-like |
| chr8_-_115167486 | 0.18 |
ENSRNOT00000033018
|
Gpr62
|
G protein-coupled receptor 62 |
| chr1_+_282134981 | 0.17 |
ENSRNOT00000036203
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr3_-_147865393 | 0.17 |
ENSRNOT00000009852
|
Sox12
|
SRY box 12 |
| chr10_-_38838272 | 0.13 |
ENSRNOT00000089495
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr6_-_11494459 | 0.12 |
ENSRNOT00000021570
|
Kcnk12
|
potassium two pore domain channel subfamily K member 12 |
| chr12_+_47193964 | 0.12 |
ENSRNOT00000001552
ENSRNOT00000039281 |
Cabp1
|
calcium binding protein 1 |
| chr2_+_210381829 | 0.11 |
ENSRNOT00000024611
|
Alx3
|
ALX homeobox 3 |
| chr3_+_175144495 | 0.10 |
ENSRNOT00000082601
ENSRNOT00000088026 |
Cdh4
|
cadherin 4 |
| chr17_-_13393243 | 0.10 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr19_-_43911057 | 0.10 |
ENSRNOT00000026017
|
Ctrb1
|
chymotrypsinogen B1 |
| chr10_-_15166457 | 0.10 |
ENSRNOT00000026676
|
Metrn
|
meteorin, glial cell differentiation regulator |
| chr9_-_82699551 | 0.10 |
ENSRNOT00000020673
|
Obsl1
|
obscurin-like 1 |
| chr1_-_277768368 | 0.09 |
ENSRNOT00000023113
|
Afap1l2
|
actin filament associated protein 1-like 2 |
| chrX_-_111942749 | 0.09 |
ENSRNOT00000087583
|
AABR07040840.1
|
|
| chr10_-_57243435 | 0.09 |
ENSRNOT00000005050
|
Chrne
|
cholinergic receptor nicotinic epsilon subunit |
| chr16_-_19918644 | 0.08 |
ENSRNOT00000083345
ENSRNOT00000023926 |
Plvap
|
plasmalemma vesicle associated protein |
| chr10_-_108196217 | 0.08 |
ENSRNOT00000075440
|
Cbx4
|
chromobox 4 |
| chr15_+_37806836 | 0.08 |
ENSRNOT00000076285
|
Il17d
|
interleukin 17D |
| chr9_+_14529218 | 0.08 |
ENSRNOT00000016893
|
Apobec2
|
apolipoprotein B mRNA editing enzyme catalytic subunit 2 |
| chr18_+_32594958 | 0.08 |
ENSRNOT00000018511
|
Spry4
|
sprouty RTK signaling antagonist 4 |
| chr5_-_79874671 | 0.08 |
ENSRNOT00000084563
|
Tnc
|
tenascin C |
| chr10_-_90312386 | 0.08 |
ENSRNOT00000028445
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr4_-_147163467 | 0.07 |
ENSRNOT00000010748
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr16_-_14300951 | 0.07 |
ENSRNOT00000017038
|
Rgr
|
retinal G protein coupled receptor |
| chr2_+_198390166 | 0.07 |
ENSRNOT00000081042
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr4_-_160605107 | 0.07 |
ENSRNOT00000090725
|
Tspan9
|
tetraspanin 9 |
| chrX_-_159841072 | 0.07 |
ENSRNOT00000001158
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr2_-_210782746 | 0.07 |
ENSRNOT00000025939
ENSRNOT00000081501 |
Gstm7
|
glutathione S-transferase, mu 7 |
| chr17_-_44793927 | 0.07 |
ENSRNOT00000086309
|
Hist1h2bo
|
histone cluster 1 H2B family member o |
| chr10_-_18063391 | 0.07 |
ENSRNOT00000072297
|
Fgf18
|
fibroblast growth factor 18 |
| chr13_+_52662996 | 0.07 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr1_+_220353356 | 0.07 |
ENSRNOT00000080189
|
Cd248
|
CD248 molecule |
| chr4_-_100099517 | 0.07 |
ENSRNOT00000014277
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr1_+_238222521 | 0.07 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr13_-_91020112 | 0.06 |
ENSRNOT00000030449
|
Dusp23
|
dual specificity phosphatase 23 |
| chr20_+_13732198 | 0.06 |
ENSRNOT00000008608
|
LOC103694877
|
macrophage migration inhibitory factor |
| chr3_+_13838304 | 0.06 |
ENSRNOT00000025067
|
Hspa5
|
heat shock protein family A member 5 |
| chr1_+_222310920 | 0.06 |
ENSRNOT00000091465
|
Macrod1
|
MACRO domain containing 1 |
| chr8_-_22874637 | 0.06 |
ENSRNOT00000064551
ENSRNOT00000090424 |
Dock6
|
dedicator of cytokinesis 6 |
| chr2_-_118745766 | 0.06 |
ENSRNOT00000013858
|
Zmat3
|
zinc finger, matrin type 3 |
| chr1_+_220362064 | 0.05 |
ENSRNOT00000074361
|
LOC108348067
|
endosialin |
| chr8_-_108261021 | 0.05 |
ENSRNOT00000040710
|
LOC100912210
|
40S ribosomal protein S25-like |
| chr15_+_4554603 | 0.05 |
ENSRNOT00000075153
|
Kcnk5
|
potassium two pore domain channel subfamily K member 5 |
| chr5_-_48436531 | 0.05 |
ENSRNOT00000060914
|
Pm20d2
|
peptidase M20 domain containing 2 |
| chr15_+_33600337 | 0.05 |
ENSRNOT00000075965
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr17_+_8299131 | 0.05 |
ENSRNOT00000083687
|
Gm45623
|
predicted gene 45623 |
| chr5_+_156215417 | 0.05 |
ENSRNOT00000067616
ENSRNOT00000077742 |
Ece1
|
endothelin converting enzyme 1 |
| chr4_+_84854386 | 0.05 |
ENSRNOT00000013620
|
Mturn
|
maturin, neural progenitor differentiation regulator homolog |
| chr17_+_44758460 | 0.05 |
ENSRNOT00000089436
|
Hist1h2an
|
histone cluster 1, H2an |
| chr17_-_43627629 | 0.05 |
ENSRNOT00000022965
|
Hist1h2af
|
histone cluster 1, H2af |
| chr7_+_72924799 | 0.05 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr5_+_63781801 | 0.05 |
ENSRNOT00000008302
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr17_+_9639330 | 0.05 |
ENSRNOT00000018232
|
Dok3
|
docking protein 3 |
| chr2_-_174413236 | 0.05 |
ENSRNOT00000064588
|
Golim4
|
golgi integral membrane protein 4 |
| chr1_-_215846911 | 0.05 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr16_-_71319449 | 0.05 |
ENSRNOT00000029284
|
Fgfr1
|
Fibroblast growth factor receptor 1 |
| chr6_-_125723732 | 0.05 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr1_-_219544328 | 0.05 |
ENSRNOT00000025847
|
Grk2
|
G protein-coupled receptor kinase 2 |
| chr18_+_25163561 | 0.05 |
ENSRNOT00000042287
ENSRNOT00000049898 ENSRNOT00000017573 |
Bin1
|
bridging integrator 1 |
| chr1_+_224938801 | 0.05 |
ENSRNOT00000073052
|
1700092M07Rik
|
RIKEN cDNA 1700092M07 gene |
| chr7_-_140580783 | 0.05 |
ENSRNOT00000087327
|
Dhh
|
desert hedgehog |
| chr1_-_15301998 | 0.05 |
ENSRNOT00000016400
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr16_-_71319052 | 0.05 |
ENSRNOT00000050980
|
Fgfr1
|
Fibroblast growth factor receptor 1 |
| chr5_-_109651730 | 0.04 |
ENSRNOT00000093032
|
Elavl2
|
ELAV like RNA binding protein 2 |
| chr12_-_38995570 | 0.04 |
ENSRNOT00000001806
|
Orai1
|
ORAI calcium release-activated calcium modulator 1 |
| chr2_+_198417619 | 0.04 |
ENSRNOT00000085945
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr9_+_49479023 | 0.04 |
ENSRNOT00000050922
ENSRNOT00000077111 |
Pou3f3
|
POU class 3 homeobox 3 |
| chr5_+_21830882 | 0.04 |
ENSRNOT00000008901
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr14_+_12218553 | 0.04 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr1_+_220446425 | 0.04 |
ENSRNOT00000027339
|
LOC100911932
|
endosialin-like |
| chr5_+_151776004 | 0.04 |
ENSRNOT00000009683
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr3_-_80933283 | 0.04 |
ENSRNOT00000007771
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr20_-_29511382 | 0.04 |
ENSRNOT00000085026
|
Ddit4
|
DNA-damage-inducible transcript 4 |
| chr15_+_33600102 | 0.04 |
ENSRNOT00000022664
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr4_-_77489535 | 0.04 |
ENSRNOT00000008728
|
Pdia4
|
protein disulfide isomerase family A, member 4 |
| chr1_-_43884267 | 0.04 |
ENSRNOT00000024418
|
Cnksr3
|
Cnksr family member 3 |
| chr1_+_86938138 | 0.04 |
ENSRNOT00000075601
|
Ccer2
|
coiled-coil glutamate-rich protein 2 |
| chr12_+_24761210 | 0.04 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr5_+_151436464 | 0.04 |
ENSRNOT00000012463
|
Map3k6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr1_-_220265772 | 0.04 |
ENSRNOT00000027119
|
Npas4
|
neuronal PAS domain protein 4 |
| chr10_+_85301875 | 0.04 |
ENSRNOT00000080935
|
Socs7
|
suppressor of cytokine signaling 7 |
| chr15_-_37383277 | 0.04 |
ENSRNOT00000011711
|
Gjb2
|
gap junction protein, beta 2 |
| chr1_+_221644867 | 0.04 |
ENSRNOT00000054830
|
Ehd1
|
EH-domain containing 1 |
| chr1_+_80321585 | 0.04 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr4_+_94696965 | 0.04 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chrX_+_82143789 | 0.04 |
ENSRNOT00000003724
|
Pou3f4
|
POU class 3 homeobox 4 |
| chr19_+_54060622 | 0.04 |
ENSRNOT00000023607
|
Gse1
|
Gse1 coiled-coil protein |
| chr5_-_144274981 | 0.04 |
ENSRNOT00000065871
|
Map7d1
|
MAP7 domain containing 1 |
| chr2_-_172361779 | 0.04 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr11_+_45888221 | 0.04 |
ENSRNOT00000071932
ENSRNOT00000066008 ENSRNOT00000060802 |
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr9_+_10339075 | 0.03 |
ENSRNOT00000073402
|
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr10_+_86860685 | 0.03 |
ENSRNOT00000084644
|
Rara
|
retinoic acid receptor, alpha |
| chr11_+_46009322 | 0.03 |
ENSRNOT00000002233
|
Tmem45a
|
transmembrane protein 45A |
| chr5_-_59025631 | 0.03 |
ENSRNOT00000049000
ENSRNOT00000022801 |
Tpm2
|
tropomyosin 2, beta |
| chr6_+_6427657 | 0.03 |
ENSRNOT00000005664
|
Pkdcc
|
protein kinase domain containing, cytoplasmic |
| chrX_+_105239840 | 0.03 |
ENSRNOT00000039864
|
Drp2
|
dystrophin related protein 2 |
| chr6_-_125723944 | 0.03 |
ENSRNOT00000007004
|
Fbln5
|
fibulin 5 |
| chr1_+_171592797 | 0.03 |
ENSRNOT00000026607
|
Syt9
|
synaptotagmin 9 |
| chr6_+_98284170 | 0.03 |
ENSRNOT00000031979
|
Rhoj
|
ras homolog family member J |
| chr1_-_265298797 | 0.03 |
ENSRNOT00000023137
|
Poll
|
DNA polymerase lambda |
| chr15_+_2374582 | 0.03 |
ENSRNOT00000019644
|
Zfp503
|
zinc finger protein 503 |
| chr18_-_14016713 | 0.03 |
ENSRNOT00000041125
|
Nol4
|
nucleolar protein 4 |
| chr10_-_89374516 | 0.03 |
ENSRNOT00000028084
|
Vat1
|
vesicle amine transport 1 |
| chr2_-_257376756 | 0.03 |
ENSRNOT00000065811
|
Gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr4_+_88328061 | 0.03 |
ENSRNOT00000084775
|
Vom1r87
|
vomeronasal 1 receptor 87 |
| chr13_-_89606326 | 0.03 |
ENSRNOT00000029179
|
Fcer1g
|
Fc fragment of IgE receptor Ig |
| chr1_+_85213652 | 0.03 |
ENSRNOT00000092044
|
Nccrp1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr10_-_82197848 | 0.03 |
ENSRNOT00000081307
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr1_+_82151669 | 0.03 |
ENSRNOT00000091357
|
Cic
|
capicua transcriptional repressor |
| chr5_+_154522119 | 0.03 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr6_+_58467254 | 0.03 |
ENSRNOT00000065396
|
Etv1
|
ets variant 1 |
| chr7_+_130532435 | 0.03 |
ENSRNOT00000092672
ENSRNOT00000092535 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr9_+_111028824 | 0.03 |
ENSRNOT00000041418
ENSRNOT00000056457 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr12_+_22165486 | 0.03 |
ENSRNOT00000001890
|
Mospd3
|
motile sperm domain containing 3 |
| chr13_-_111581018 | 0.03 |
ENSRNOT00000083072
ENSRNOT00000077981 |
Sertad4
|
SERTA domain containing 4 |
| chr12_-_12025549 | 0.03 |
ENSRNOT00000001331
|
Nptx2
|
neuronal pentraxin 2 |
| chr4_-_116278615 | 0.03 |
ENSRNOT00000020505
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr10_-_4910305 | 0.03 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr1_+_220416018 | 0.03 |
ENSRNOT00000027233
|
B4gat1
|
beta-1,4-glucuronyltransferase 1 |
| chr6_-_104290579 | 0.03 |
ENSRNOT00000066014
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr7_-_3342491 | 0.03 |
ENSRNOT00000081756
|
Rdh5
|
retinol dehydrogenase 5 |
| chr10_+_106812739 | 0.03 |
ENSRNOT00000074225
|
Syngr2
|
synaptogyrin 2 |
| chr13_-_48284408 | 0.03 |
ENSRNOT00000085967
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr13_+_95589668 | 0.03 |
ENSRNOT00000005849
|
Zfp238
|
zinc finger protein 238 |
| chr8_+_111495331 | 0.03 |
ENSRNOT00000091275
ENSRNOT00000012162 |
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chr7_-_3315856 | 0.03 |
ENSRNOT00000010035
|
Gdf11
|
growth differentiation factor 11 |
| chr14_-_8548310 | 0.02 |
ENSRNOT00000092436
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr17_-_43807540 | 0.02 |
ENSRNOT00000074763
|
LOC684762
|
similar to CG31613-PA |
| chr8_-_119012671 | 0.02 |
ENSRNOT00000028435
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr6_-_102196138 | 0.02 |
ENSRNOT00000014132
|
Tmem229b
|
transmembrane protein 229B |
| chr9_-_18371856 | 0.02 |
ENSRNOT00000027293
|
Supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr4_-_157263890 | 0.02 |
ENSRNOT00000065416
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr3_-_138116664 | 0.02 |
ENSRNOT00000055602
|
Rrbp1
|
ribosome binding protein 1 |
| chr11_+_82194657 | 0.02 |
ENSRNOT00000002445
|
Etv5
|
ets variant 5 |
| chr6_+_35314444 | 0.02 |
ENSRNOT00000005666
|
Osr1
|
odd-skipped related transciption factor 1 |
| chr3_+_11207542 | 0.02 |
ENSRNOT00000013546
|
Prrc2b
|
proline-rich coiled-coil 2B |
| chr3_-_2534375 | 0.02 |
ENSRNOT00000037725
|
Grin1
|
glutamate ionotropic receptor NMDA type subunit 1 |
| chr5_-_113532878 | 0.02 |
ENSRNOT00000010173
|
Caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr14_+_13192347 | 0.02 |
ENSRNOT00000000092
|
Antxr2
|
anthrax toxin receptor 2 |
| chr19_-_26053762 | 0.02 |
ENSRNOT00000004646
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr15_+_23792931 | 0.02 |
ENSRNOT00000092091
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr12_-_25638797 | 0.02 |
ENSRNOT00000002033
|
Gatsl2
|
GATS protein-like 2 |
| chr1_+_170471238 | 0.02 |
ENSRNOT00000076961
ENSRNOT00000075597 ENSRNOT00000076631 ENSRNOT00000076783 |
Timm10b
Dnhd1
|
translocase of inner mitochondrial membrane 10B dynein heavy chain domain 1 |
| chr1_-_166912524 | 0.02 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr1_-_174350196 | 0.02 |
ENSRNOT00000055158
|
LOC499240
|
similar to predicted gene ICRFP703B1614Q5.5 |
| chr9_+_84689150 | 0.02 |
ENSRNOT00000072242
|
Kcne4
|
potassium voltage-gated channel subfamily E regulatory subunit 4 |
| chr12_-_36398206 | 0.02 |
ENSRNOT00000090944
ENSRNOT00000058492 |
Tmem132b
|
transmembrane protein 132B |
| chr7_-_77372398 | 0.02 |
ENSRNOT00000079644
ENSRNOT00000007999 |
Azin1
|
antizyme inhibitor 1 |
| chr10_+_89339029 | 0.02 |
ENSRNOT00000031208
|
Rundc1
|
RUN domain containing 1 |
| chr1_+_157646173 | 0.02 |
ENSRNOT00000013851
|
Rab30
|
RAB30, member RAS oncogene family |
| chr9_+_82556573 | 0.02 |
ENSRNOT00000026860
|
Des
|
desmin |
| chr7_+_70980422 | 0.02 |
ENSRNOT00000077912
|
Rdh16
|
retinol dehydrogenase 16 (all-trans) |
| chr1_+_101214593 | 0.02 |
ENSRNOT00000028086
|
Tead2
|
TEA domain transcription factor 2 |
| chr7_+_120153184 | 0.02 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chr11_+_46714355 | 0.02 |
ENSRNOT00000073404
|
Tmem45al
|
transmembrane protein 45A-like |
| chr12_+_22641104 | 0.02 |
ENSRNOT00000001916
|
Serpine1
|
serpin family E member 1 |
| chr4_+_96831880 | 0.02 |
ENSRNOT00000068400
|
RSA-14-44
|
RSA-14-44 protein |
| chr10_+_54126486 | 0.02 |
ENSRNOT00000078820
|
Gas7
|
growth arrest specific 7 |
| chr4_-_159697207 | 0.02 |
ENSRNOT00000086440
|
Ccnd2
|
cyclin D2 |
| chr18_+_60392376 | 0.02 |
ENSRNOT00000023890
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr4_+_129574264 | 0.02 |
ENSRNOT00000010185
|
Arl6ip5
|
ADP-ribosylation factor like GTPase 6 interacting protein 5 |
| chr10_+_109446444 | 0.02 |
ENSRNOT00000066736
|
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chrX_+_80213332 | 0.02 |
ENSRNOT00000042827
|
Sh3bgrl
|
SH3 domain binding glutamate-rich protein like |
| chr8_-_116993193 | 0.02 |
ENSRNOT00000026327
|
Dag1
|
dystroglycan 1 |
| chr10_-_40122915 | 0.02 |
ENSRNOT00000063891
|
Cdc42se2
|
CDC42 small effector 2 |
| chr1_-_101054851 | 0.02 |
ENSRNOT00000027849
|
Prrg2
|
proline rich and Gla domain 2 |
| chr5_-_88612626 | 0.02 |
ENSRNOT00000089560
|
Tle1
|
transducin like enhancer of split 1 |
| chr17_-_9565249 | 0.02 |
ENSRNOT00000036140
|
Tmed9
|
transmembrane p24 trafficking protein 9 |
| chr1_+_13595295 | 0.02 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr7_-_126461658 | 0.02 |
ENSRNOT00000081032
|
Wnt7b
|
wingless-type MMTV integration site family, member 7B |
| chr16_-_59721227 | 0.02 |
ENSRNOT00000015166
|
Prag1
|
PEAK1 related kinase activating pseudokinase 1 |
| chr9_-_82419288 | 0.02 |
ENSRNOT00000004797
|
Tuba4a
|
tubulin, alpha 4A |
| chr11_-_65209268 | 0.02 |
ENSRNOT00000077612
|
Gsk3b
|
glycogen synthase kinase 3 beta |
| chr3_+_5624506 | 0.02 |
ENSRNOT00000036995
|
Adamtsl2
|
ADAMTS-like 2 |
| chr2_-_186245163 | 0.02 |
ENSRNOT00000089339
|
Dclk2
|
doublecortin-like kinase 2 |
| chr19_-_56772904 | 0.02 |
ENSRNOT00000024232
|
Abcb10
|
ATP binding cassette subfamily B member 10 |
| chr1_+_198214797 | 0.02 |
ENSRNOT00000068543
|
Tbx6
|
T-box 6 |
| chr10_-_15928169 | 0.02 |
ENSRNOT00000028069
|
Nsg2
|
neuron specific gene family member 2 |
| chr2_-_143104412 | 0.02 |
ENSRNOT00000058116
|
Ufm1
|
ubiquitin-fold modifier 1 |
| chr14_-_86395120 | 0.02 |
ENSRNOT00000006802
ENSRNOT00000078572 |
Tmed4
|
transmembrane p24 trafficking protein 4 |
| chr1_+_15412603 | 0.02 |
ENSRNOT00000051496
ENSRNOT00000067070 |
Map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr11_-_90406797 | 0.02 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
| chr4_-_7252950 | 0.02 |
ENSRNOT00000084777
ENSRNOT00000087120 |
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr1_-_89007041 | 0.02 |
ENSRNOT00000032363
|
Proser3
|
proline and serine rich 3 |
| chr18_+_73256085 | 0.02 |
ENSRNOT00000080660
|
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr1_+_214317682 | 0.02 |
ENSRNOT00000032904
|
Tmem80
|
transmembrane protein 80 |
| chr1_+_220322940 | 0.02 |
ENSRNOT00000074972
|
LOC108348085
|
beta-1,4-glucuronyltransferase 1 |
| chr1_+_277355619 | 0.02 |
ENSRNOT00000022788
|
Nhlrc2
|
NHL repeat containing 2 |
| chr14_-_86739335 | 0.01 |
ENSRNOT00000078282
|
Purb
|
purine rich element binding protein B |
| chr4_+_8066907 | 0.01 |
ENSRNOT00000080811
|
Srpk2
|
SRSF protein kinase 2 |
| chr1_-_214971929 | 0.01 |
ENSRNOT00000027195
|
Mob2
|
MOB kinase activator 2 |
| chr14_+_66598259 | 0.01 |
ENSRNOT00000049743
|
Kcnip4
|
potassium voltage-gated channel interacting protein 4 |
| chr3_-_172537877 | 0.01 |
ENSRNOT00000072069
|
Ctsz
|
cathepsin Z |
| chr1_-_277355075 | 0.01 |
ENSRNOT00000036321
|
Dclre1a
|
DNA cross-link repair 1A |
| chr5_+_48313599 | 0.01 |
ENSRNOT00000081825
|
Gabrr2
|
gamma-aminobutyric acid type A receptor rho 2 subunit |
| chr9_+_9961021 | 0.01 |
ENSRNOT00000075767
|
Tubb4a
|
tubulin, beta 4A class IVa |
| chr10_+_74959285 | 0.01 |
ENSRNOT00000010296
|
Rnf43
|
ring finger protein 43 |
| chr7_-_130101858 | 0.01 |
ENSRNOT00000007954
|
Tubgcp6
|
tubulin, gamma complex associated protein 6 |
| chr9_+_82053581 | 0.01 |
ENSRNOT00000086375
|
Wnt10a
|
wingless-type MMTV integration site family, member 10A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.2 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.0 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0016554 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine to uridine editing(GO:0016554) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.0 | 0.1 | GO:0071799 | bud outgrowth involved in lung branching(GO:0060447) response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.1 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 0.0 | 0.0 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.0 | GO:0072218 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |