Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Tcf21_Msc
Z-value: 0.93
Transcription factors associated with Tcf21_Msc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
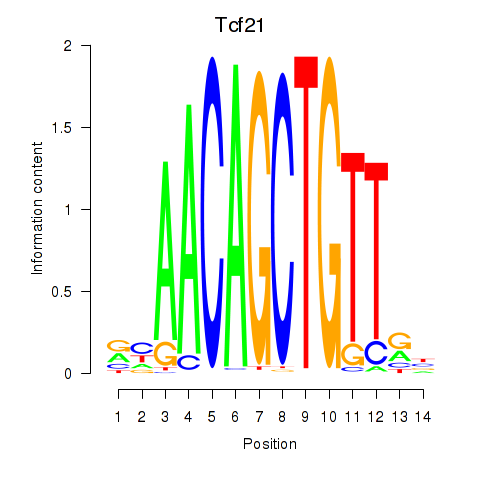
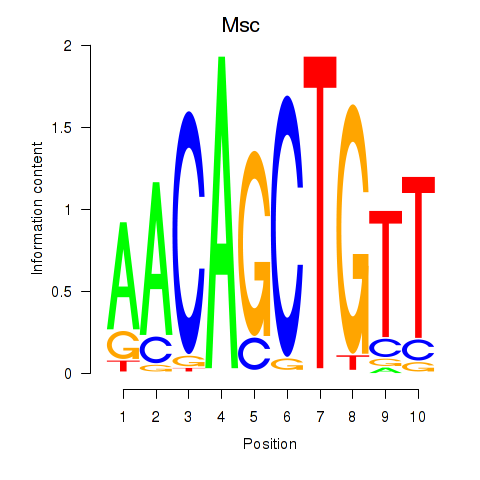
Tcf21
|
ENSRNOG00000016700 | transcription factor 21 |
|
Msc
|
ENSRNOG00000007540 | musculin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf21 | rn6_v1_chr1_+_23906717_23906717 | -0.72 | 1.7e-01 | Click! |
| Msc | rn6_v1_chr5_+_3955227_3955227 | -0.39 | 5.2e-01 | Click! |
Activity profile of Tcf21_Msc motif
Sorted Z-values of Tcf21_Msc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_9226125 | 0.56 |
ENSRNOT00000088047
|
Wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr10_-_64642292 | 0.53 |
ENSRNOT00000084670
|
Abr
|
active BCR-related |
| chr14_-_92077088 | 0.48 |
ENSRNOT00000085175
|
Grb10
|
growth factor receptor bound protein 10 |
| chr5_+_90800082 | 0.45 |
ENSRNOT00000093696
ENSRNOT00000093206 |
Kdm4c
|
lysine demethylase 4C |
| chr10_+_85301875 | 0.42 |
ENSRNOT00000080935
|
Socs7
|
suppressor of cytokine signaling 7 |
| chr10_+_71278650 | 0.40 |
ENSRNOT00000092020
|
Synrg
|
synergin, gamma |
| chr11_+_38035450 | 0.40 |
ENSRNOT00000083067
|
Mx2
|
MX dynamin like GTPase 2 |
| chr1_+_213583606 | 0.39 |
ENSRNOT00000088899
|
AC109844.1
|
|
| chr1_+_278557792 | 0.39 |
ENSRNOT00000023414
|
Atrnl1
|
attractin like 1 |
| chr6_-_138401621 | 0.36 |
ENSRNOT00000043210
|
LOC103693608
|
integral membrane protein DGCR2/IDD-like |
| chr1_-_166912524 | 0.36 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr2_-_178521038 | 0.35 |
ENSRNOT00000033107
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr17_-_51912496 | 0.32 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr5_-_168734296 | 0.32 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr2_+_196594303 | 0.31 |
ENSRNOT00000064442
ENSRNOT00000044738 |
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr17_-_10208360 | 0.31 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr20_+_3351303 | 0.30 |
ENSRNOT00000080419
ENSRNOT00000001065 ENSRNOT00000086503 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr3_+_72329967 | 0.30 |
ENSRNOT00000090256
|
Slc43a3
|
solute carrier family 43, member 3 |
| chr9_+_94425252 | 0.28 |
ENSRNOT00000064965
ENSRNOT00000076099 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr1_-_199439210 | 0.27 |
ENSRNOT00000026699
|
Pycard
|
PYD and CARD domain containing |
| chr13_+_50196042 | 0.27 |
ENSRNOT00000090858
|
Zc3h11a
|
zinc finger CCCH-type containing 11A |
| chr5_+_167141875 | 0.27 |
ENSRNOT00000089314
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr7_-_145450233 | 0.25 |
ENSRNOT00000092974
ENSRNOT00000021523 |
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr11_-_90406797 | 0.25 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
| chr12_-_25638797 | 0.25 |
ENSRNOT00000002033
|
Gatsl2
|
GATS protein-like 2 |
| chr1_-_212622537 | 0.25 |
ENSRNOT00000025609
|
Sprn
|
shadow of prion protein homolog (zebrafish) |
| chr18_-_55891710 | 0.25 |
ENSRNOT00000064686
|
Synpo
|
synaptopodin |
| chrX_-_26376467 | 0.25 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr6_+_26537707 | 0.24 |
ENSRNOT00000050102
|
Zfp513
|
zinc finger protein 513 |
| chrX_-_39956841 | 0.24 |
ENSRNOT00000050708
|
Klhl34
|
kelch-like family member 34 |
| chr4_+_157538303 | 0.24 |
ENSRNOT00000086418
|
Zfp384
|
zinc finger protein 384 |
| chr12_-_47987255 | 0.23 |
ENSRNOT00000074000
|
Ube3b
|
ubiquitin protein ligase E3B |
| chr5_-_154363329 | 0.23 |
ENSRNOT00000064596
|
Eloa
|
elongin A |
| chr9_-_80166807 | 0.23 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr1_-_167911961 | 0.23 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr4_+_157126935 | 0.23 |
ENSRNOT00000056051
|
C1r
|
complement C1r |
| chr1_+_192233910 | 0.23 |
ENSRNOT00000016418
ENSRNOT00000016442 |
Prkcb
|
protein kinase C, beta |
| chr3_-_112876773 | 0.22 |
ENSRNOT00000015086
|
Ubr1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr20_+_14101659 | 0.22 |
ENSRNOT00000072696
|
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr8_+_116343096 | 0.22 |
ENSRNOT00000022092
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr4_-_145147397 | 0.22 |
ENSRNOT00000010347
|
Lhfpl4
|
lipoma HMGIC fusion partner-like 4 |
| chr2_-_261402880 | 0.21 |
ENSRNOT00000052396
ENSRNOT00000077839 ENSRNOT00000088561 |
Fpgt
|
fucose-1-phosphate guanylyltransferase |
| chr8_+_53295222 | 0.21 |
ENSRNOT00000009724
ENSRNOT00000067420 |
Usp28
|
ubiquitin specific peptidase 28 |
| chr11_+_38035611 | 0.21 |
ENSRNOT00000087603
ENSRNOT00000002695 |
Mx2
|
MX dynamin like GTPase 2 |
| chr9_+_111028824 | 0.21 |
ENSRNOT00000041418
ENSRNOT00000056457 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr7_-_130128589 | 0.21 |
ENSRNOT00000079777
ENSRNOT00000009325 |
Mapk11
|
mitogen-activated protein kinase 11 |
| chr7_+_73588163 | 0.21 |
ENSRNOT00000015707
|
Kcns2
|
potassium voltage-gated channel, modifier subfamily S, member 2 |
| chr10_+_75055020 | 0.21 |
ENSRNOT00000010755
|
Tspoap1
|
TSPO associated protein 1 |
| chr12_+_660011 | 0.21 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr1_-_215846911 | 0.20 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr14_-_78377825 | 0.20 |
ENSRNOT00000068104
|
AABR07015812.1
|
|
| chr19_-_34752695 | 0.20 |
ENSRNOT00000052018
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr5_+_5616483 | 0.19 |
ENSRNOT00000011026
|
Ncoa2
|
nuclear receptor coactivator 2 |
| chr5_+_122390522 | 0.19 |
ENSRNOT00000064567
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr20_+_3827367 | 0.19 |
ENSRNOT00000079967
|
Rxrb
|
retinoid X receptor beta |
| chr11_+_84745904 | 0.19 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr14_-_114649173 | 0.19 |
ENSRNOT00000083528
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr7_-_142300382 | 0.19 |
ENSRNOT00000048262
|
Bin2
|
bridging integrator 2 |
| chr7_+_12146642 | 0.19 |
ENSRNOT00000085899
|
Tcf3
|
transcription factor 3 |
| chr8_+_62925357 | 0.18 |
ENSRNOT00000011074
ENSRNOT00000090588 |
Stra6
|
stimulated by retinoic acid 6 |
| chr12_+_36734372 | 0.18 |
ENSRNOT00000001301
|
Scarb1
|
scavenger receptor class B, member 1 |
| chr11_-_82664554 | 0.18 |
ENSRNOT00000002425
ENSRNOT00000087105 |
Senp2
|
Sumo1/sentrin/SMT3 specific peptidase 2 |
| chr14_+_12218553 | 0.17 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr17_-_78910671 | 0.17 |
ENSRNOT00000064609
|
Acbd7
|
acyl-CoA binding domain containing 7 |
| chr5_-_153790157 | 0.17 |
ENSRNOT00000025051
|
Rcan3
|
RCAN family member 3 |
| chr5_+_63781801 | 0.17 |
ENSRNOT00000008302
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr18_+_56180089 | 0.17 |
ENSRNOT00000030966
|
Arsi
|
arylsulfatase family, member I |
| chr11_-_43022565 | 0.16 |
ENSRNOT00000002285
|
Riox2
|
ribosomal oxygenase 2 |
| chr2_+_243840134 | 0.16 |
ENSRNOT00000093355
|
Eif4e
|
eukaryotic translation initiation factor 4E |
| chr10_-_51669297 | 0.16 |
ENSRNOT00000071595
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr14_-_84106997 | 0.16 |
ENSRNOT00000065501
|
Osbp2
|
oxysterol binding protein 2 |
| chr1_-_191651628 | 0.16 |
ENSRNOT00000055048
|
Usp31
|
ubiquitin specific peptidase 31 |
| chr2_-_88135410 | 0.16 |
ENSRNOT00000014180
|
Car3
|
carbonic anhydrase 3 |
| chr12_-_37185548 | 0.16 |
ENSRNOT00000001338
|
LOC102556092
|
uncharacterized LOC102556092 |
| chr7_-_117712213 | 0.16 |
ENSRNOT00000092930
|
Cyhr1
|
cysteine and histidine rich 1 |
| chr1_+_266380973 | 0.15 |
ENSRNOT00000080509
|
Wbp1l
|
WW domain binding protein 1-like |
| chr3_-_61494778 | 0.15 |
ENSRNOT00000068018
|
Lnpk
|
lunapark, ER junction formation factor |
| chr2_-_118989127 | 0.15 |
ENSRNOT00000014871
|
Gnb4
|
G protein subunit beta 4 |
| chr5_-_137014375 | 0.15 |
ENSRNOT00000066165
ENSRNOT00000091866 |
Kdm4a
|
lysine demethylase 4A |
| chr5_+_145138697 | 0.15 |
ENSRNOT00000019135
|
Zmym6
|
zinc finger MYM-type containing 6 |
| chr7_-_11777503 | 0.14 |
ENSRNOT00000026220
|
Amh
|
anti-Mullerian hormone |
| chr6_-_127248372 | 0.14 |
ENSRNOT00000085517
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr5_-_158439078 | 0.14 |
ENSRNOT00000025517
|
Klhdc7a
|
kelch domain containing 7A |
| chr4_+_21872856 | 0.14 |
ENSRNOT00000044810
|
Dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chr1_-_277768368 | 0.14 |
ENSRNOT00000023113
|
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr13_+_50434702 | 0.14 |
ENSRNOT00000032244
|
Sox13
|
SRY box 13 |
| chr1_+_13595295 | 0.14 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr5_-_58469399 | 0.14 |
ENSRNOT00000013213
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr8_+_58120179 | 0.14 |
ENSRNOT00000049076
ENSRNOT00000090114 |
Npat
|
nuclear protein, co-activator of histone transcription |
| chr3_-_7422738 | 0.14 |
ENSRNOT00000088339
|
Gtf3c4
|
general transcription factor IIIC subunit 4 |
| chr2_-_45518502 | 0.14 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr20_+_28572242 | 0.14 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr3_+_172856733 | 0.14 |
ENSRNOT00000009826
|
Edn3
|
endothelin 3 |
| chr9_-_65307995 | 0.13 |
ENSRNOT00000030934
|
Clk1
|
CDC-like kinase 1 |
| chr19_+_45938915 | 0.13 |
ENSRNOT00000065508
|
Mon1b
|
MON1 homolog B, secretory trafficking associated |
| chr3_-_12007570 | 0.13 |
ENSRNOT00000060186
|
Lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr16_-_79973735 | 0.13 |
ENSRNOT00000057845
ENSRNOT00000086896 |
Dlgap2
|
DLG associated protein 2 |
| chr5_-_136965191 | 0.13 |
ENSRNOT00000056842
|
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr15_+_4554603 | 0.13 |
ENSRNOT00000075153
|
Kcnk5
|
potassium two pore domain channel subfamily K member 5 |
| chr8_+_68526093 | 0.13 |
ENSRNOT00000011385
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chr3_-_110324084 | 0.13 |
ENSRNOT00000010381
|
Bmf
|
Bcl2 modifying factor |
| chr1_-_278042312 | 0.13 |
ENSRNOT00000018693
|
Ablim1
|
actin-binding LIM protein 1 |
| chr2_+_210381829 | 0.13 |
ENSRNOT00000024611
|
Alx3
|
ALX homeobox 3 |
| chr15_-_37383277 | 0.13 |
ENSRNOT00000011711
|
Gjb2
|
gap junction protein, beta 2 |
| chr15_-_33725188 | 0.13 |
ENSRNOT00000083941
|
Zfhx2
|
zinc finger homeobox 2 |
| chr5_+_48274477 | 0.13 |
ENSRNOT00000075992
ENSRNOT00000041271 |
Ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr16_-_14348046 | 0.13 |
ENSRNOT00000018630
|
Cdhr1
|
cadherin-related family member 1 |
| chr3_-_171286413 | 0.13 |
ENSRNOT00000008365
ENSRNOT00000081036 |
Zbp1
|
Z-DNA binding protein 1 |
| chr8_-_58119623 | 0.13 |
ENSRNOT00000050555
|
Atm
|
ATM serine/threonine kinase |
| chr18_+_14471213 | 0.12 |
ENSRNOT00000045224
|
Dtna
|
dystrobrevin, alpha |
| chr10_-_74679858 | 0.12 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr13_+_89805962 | 0.12 |
ENSRNOT00000074035
|
Tstd1
|
thiosulfate sulfurtransferase like domain containing 1 |
| chr15_-_42898150 | 0.12 |
ENSRNOT00000030036
|
Ptk2b
|
protein tyrosine kinase 2 beta |
| chr13_-_48284408 | 0.12 |
ENSRNOT00000085967
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr15_-_106678918 | 0.12 |
ENSRNOT00000034723
|
Stk24
|
serine/threonine kinase 24 |
| chr4_+_25538497 | 0.12 |
ENSRNOT00000009264
|
Cfap69
|
cilia and flagella associated protein 69 |
| chr12_-_22126350 | 0.12 |
ENSRNOT00000076328
|
Sap25
|
Sin3A-associated protein 25 |
| chr1_+_80321585 | 0.12 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr15_-_55209342 | 0.12 |
ENSRNOT00000021752
|
Rb1
|
RB transcriptional corepressor 1 |
| chrX_+_71528988 | 0.12 |
ENSRNOT00000075963
|
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr3_+_110723481 | 0.12 |
ENSRNOT00000013878
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr12_+_48577905 | 0.12 |
ENSRNOT00000000888
|
Selplg
|
selectin P ligand |
| chr18_+_44737154 | 0.12 |
ENSRNOT00000021972
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chr14_+_81488008 | 0.12 |
ENSRNOT00000018477
|
Tnip2
|
TNFAIP3 interacting protein 2 |
| chr17_+_77195247 | 0.12 |
ENSRNOT00000081940
|
Optn
|
optineurin |
| chr8_-_116531784 | 0.12 |
ENSRNOT00000024529
|
Rbm5
|
RNA binding motif protein 5 |
| chr10_+_62191656 | 0.11 |
ENSRNOT00000029866
|
Smyd4
|
SET and MYND domain containing 4 |
| chr4_-_22425381 | 0.11 |
ENSRNOT00000011166
ENSRNOT00000037712 |
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr7_-_12275609 | 0.11 |
ENSRNOT00000086061
|
Apc2
|
APC2, WNT signaling pathway regulator |
| chr17_+_9596957 | 0.11 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr6_+_79254339 | 0.11 |
ENSRNOT00000073970
|
Sstr1
|
somatostatin receptor 1 |
| chr18_+_30387937 | 0.11 |
ENSRNOT00000027210
|
Pcdhb4
|
protocadherin beta 4 |
| chr4_+_68011932 | 0.11 |
ENSRNOT00000073638
|
Tmem178b
|
transmembrane protein 178B |
| chr4_+_168599331 | 0.11 |
ENSRNOT00000086719
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr14_+_69800156 | 0.11 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr12_-_41993957 | 0.11 |
ENSRNOT00000001891
|
Rbm19
|
RNA binding motif protein 19 |
| chr2_-_40386669 | 0.11 |
ENSRNOT00000014074
|
Elovl7
|
ELOVL fatty acid elongase 7 |
| chr14_+_86652365 | 0.11 |
ENSRNOT00000077428
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr19_+_755460 | 0.11 |
ENSRNOT00000076560
|
Dync1li2
|
dynein, cytoplasmic 1 light intermediate chain 2 |
| chr8_-_114013623 | 0.11 |
ENSRNOT00000018175
|
Atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr14_-_28856214 | 0.11 |
ENSRNOT00000044659
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr7_-_140600818 | 0.11 |
ENSRNOT00000082375
|
Lmbr1l
|
limb development membrane protein 1-like |
| chr9_+_65331290 | 0.11 |
ENSRNOT00000058667
ENSRNOT00000018014 |
Nif3l1
|
NGG1 interacting factor 3 like 1 |
| chr14_-_45797887 | 0.11 |
ENSRNOT00000029209
|
Tbc1d1
|
TBC1 domain family member 1 |
| chr9_+_47281961 | 0.11 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chr3_+_5555807 | 0.11 |
ENSRNOT00000007677
|
Cacfd1
|
calcium channel flower domain containing 1 |
| chr10_+_54156649 | 0.11 |
ENSRNOT00000074718
|
Gas7
|
growth arrest specific 7 |
| chr7_-_97977135 | 0.10 |
ENSRNOT00000008785
|
Wdyhv1
|
WDYHV motif containing 1 |
| chr2_+_77868412 | 0.10 |
ENSRNOT00000065897
ENSRNOT00000014022 |
Myo10
|
myosin X |
| chr2_+_197720259 | 0.10 |
ENSRNOT00000070919
|
Golph3l
|
golgi phosphoprotein 3-like |
| chr7_+_137155481 | 0.10 |
ENSRNOT00000089895
|
Ano6
|
anoctamin 6 |
| chr1_+_85112834 | 0.10 |
ENSRNOT00000026107
|
Dyrk1b
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr10_-_43512287 | 0.10 |
ENSRNOT00000036481
|
Faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr1_-_214971929 | 0.10 |
ENSRNOT00000027195
|
Mob2
|
MOB kinase activator 2 |
| chr12_-_22127021 | 0.10 |
ENSRNOT00000076261
|
Sap25
|
Sin3A-associated protein 25 |
| chr19_-_41433346 | 0.10 |
ENSRNOT00000022952
|
Cmtr2
|
cap methyltransferase 2 |
| chr18_+_27424328 | 0.10 |
ENSRNOT00000033784
|
Kif20a
|
kinesin family member 20A |
| chrX_+_156812064 | 0.10 |
ENSRNOT00000077142
|
Hcfc1
|
host cell factor C1 |
| chr1_+_80000165 | 0.10 |
ENSRNOT00000084912
|
Six5
|
SIX homeobox 5 |
| chr4_+_110699557 | 0.10 |
ENSRNOT00000030588
ENSRNOT00000092261 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr4_-_159697207 | 0.10 |
ENSRNOT00000086440
|
Ccnd2
|
cyclin D2 |
| chr8_-_96132634 | 0.10 |
ENSRNOT00000041655
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr6_-_15191660 | 0.10 |
ENSRNOT00000092654
|
Nrxn1
|
neurexin 1 |
| chr13_+_50873605 | 0.10 |
ENSRNOT00000004382
|
Fmod
|
fibromodulin |
| chr1_-_222272285 | 0.10 |
ENSRNOT00000028737
|
Fermt3
|
fermitin family member 3 |
| chr2_-_210943620 | 0.10 |
ENSRNOT00000026750
|
Gpr61
|
G protein-coupled receptor 61 |
| chr1_-_252808380 | 0.10 |
ENSRNOT00000025856
|
Ch25h
|
cholesterol 25-hydroxylase |
| chr20_+_3363737 | 0.10 |
ENSRNOT00000079654
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr13_-_73558912 | 0.10 |
ENSRNOT00000093655
|
Cep350
|
centrosomal protein 350 |
| chr3_+_8547349 | 0.10 |
ENSRNOT00000079108
ENSRNOT00000091697 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr7_-_120882392 | 0.10 |
ENSRNOT00000056179
ENSRNOT00000056178 |
Fam227a
|
family with sequence similarity 227, member A |
| chr7_-_140580783 | 0.09 |
ENSRNOT00000087327
|
Dhh
|
desert hedgehog |
| chr2_-_120316357 | 0.09 |
ENSRNOT00000065469
|
Ccdc39
|
coiled-coil domain containing 39 |
| chr18_+_17043903 | 0.09 |
ENSRNOT00000068139
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr3_+_7237192 | 0.09 |
ENSRNOT00000016904
|
Tsc1
|
tuberous sclerosis 1 |
| chr3_-_110556808 | 0.09 |
ENSRNOT00000092158
ENSRNOT00000045362 |
RGD1565536
|
similar to hypothetical protein |
| chr4_-_125929002 | 0.09 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr10_+_83231238 | 0.09 |
ENSRNOT00000077625
|
Spop
|
speckle type BTB/POZ protein |
| chr1_-_90956544 | 0.09 |
ENSRNOT00000051422
|
LOC108348128
|
nesprin-4 |
| chr1_+_124625985 | 0.09 |
ENSRNOT00000021068
|
Otud7a
|
OTU deubiquitinase 7A |
| chr1_+_104635989 | 0.09 |
ENSRNOT00000078477
|
Nav2
|
neuron navigator 2 |
| chr11_-_83926524 | 0.09 |
ENSRNOT00000041777
ENSRNOT00000040029 |
Eif4g1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr17_+_27496353 | 0.09 |
ENSRNOT00000077994
ENSRNOT00000019040 ENSRNOT00000089736 |
Ssr1
|
signal sequence receptor subunit 1 |
| chr1_+_220353356 | 0.09 |
ENSRNOT00000080189
|
Cd248
|
CD248 molecule |
| chr14_+_85250042 | 0.09 |
ENSRNOT00000057407
|
Ap1b1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr4_+_127164453 | 0.09 |
ENSRNOT00000017889
|
Kbtbd8
|
kelch repeat and BTB domain containing 8 |
| chr8_+_77107536 | 0.09 |
ENSRNOT00000083255
|
Adam10
|
ADAM metallopeptidase domain 10 |
| chr3_+_176818437 | 0.09 |
ENSRNOT00000087279
|
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr10_-_47724499 | 0.09 |
ENSRNOT00000085011
|
Rnf112
|
ring finger protein 112 |
| chr16_-_19643236 | 0.09 |
ENSRNOT00000087597
|
Zfp709
|
zinc finger protein 709 |
| chr4_-_67301102 | 0.09 |
ENSRNOT00000034549
|
Dennd2a
|
DENN domain containing 2A |
| chr12_-_38504774 | 0.09 |
ENSRNOT00000011286
|
B3gnt4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr13_+_44812567 | 0.08 |
ENSRNOT00000005372
|
R3hdm1
|
R3H domain containing 1 |
| chr10_-_67285617 | 0.08 |
ENSRNOT00000019044
|
Utp6
|
UTP6 small subunit processome component |
| chr9_-_10659357 | 0.08 |
ENSRNOT00000091450
|
Kdm4b
|
lysine demethylase 4B |
| chr9_+_82571269 | 0.08 |
ENSRNOT00000026941
|
Speg
|
SPEG complex locus |
| chr2_+_185343883 | 0.08 |
ENSRNOT00000093640
|
Sh3d19
|
SH3 domain containing 19 |
| chr1_+_220362064 | 0.08 |
ENSRNOT00000074361
|
LOC108348067
|
endosialin |
| chr15_-_62200837 | 0.08 |
ENSRNOT00000017599
|
Pcdh8
|
protocadherin 8 |
| chr3_-_64554953 | 0.08 |
ENSRNOT00000067452
|
LOC102553814
|
serine/arginine repetitive matrix protein 1-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf21_Msc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.4 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.1 | 0.6 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.4 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 0.3 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) |
| 0.1 | 0.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.3 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.2 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.1 | 0.7 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.2 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 0.5 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.1 | 0.6 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.1 | 0.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 0.2 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.1 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.2 | GO:0046864 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.1 | 0.2 | GO:0032497 | lipopolysaccharide transport(GO:0015920) detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.2 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 0.3 | GO:0015755 | fructose transport(GO:0015755) |
| 0.1 | 0.2 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.2 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.2 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.2 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.1 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.2 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.2 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:1905230 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0019046 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.0 | 0.3 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0071283 | cellular response to iron(III) ion(GO:0071283) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.0 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.4 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0060383 | regulation of DNA strand elongation(GO:0060382) positive regulation of DNA strand elongation(GO:0060383) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0072054 | renal outer medulla development(GO:0072054) |
| 0.0 | 0.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.1 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:0060558 | regulation of calcidiol 1-monooxygenase activity(GO:0060558) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.0 | GO:1904404 | traversing start control point of mitotic cell cycle(GO:0007089) response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.2 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.1 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.0 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 | 0.0 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.2 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) D-amino acid metabolic process(GO:0046416) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.3 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.1 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.3 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.3 | GO:0097169 | NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0031308 | intrinsic component of nuclear outer membrane(GO:0031308) integral component of nuclear outer membrane(GO:0031309) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.3 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.1 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.2 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 0.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.2 | GO:0016842 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.4 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.3 | GO:0070061 | fructose binding(GO:0070061) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.2 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.0 | 0.0 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |