Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
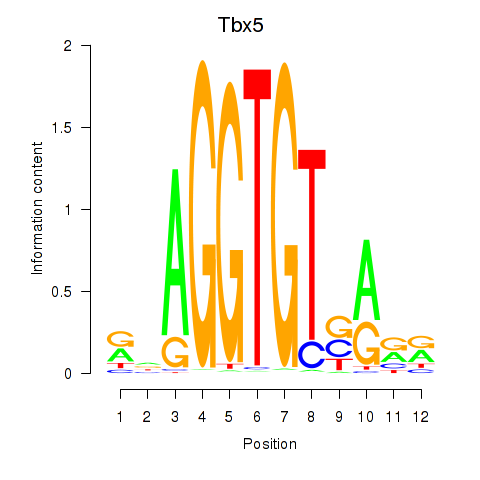
Results for Tbx5
Z-value: 0.52
Transcription factors associated with Tbx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx5
|
ENSRNOG00000001399 | T-box 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx5 | rn6_v1_chr12_+_42097626_42097626 | 0.34 | 5.7e-01 | Click! |
Activity profile of Tbx5 motif
Sorted Z-values of Tbx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_15274917 | 0.26 |
ENSRNOT00000019650
|
Pgc
|
progastricsin |
| chr16_-_8564379 | 0.20 |
ENSRNOT00000060975
|
LOC680885
|
hypothetical protein LOC680885 |
| chr5_+_164808323 | 0.20 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr6_+_135856218 | 0.20 |
ENSRNOT00000066715
|
RGD1560608
|
similar to novel protein |
| chr2_-_30127269 | 0.20 |
ENSRNOT00000023869
|
Cartpt
|
CART prepropeptide |
| chr1_+_80777014 | 0.18 |
ENSRNOT00000079758
|
Gm19345
|
predicted gene, 19345 |
| chr8_+_89369417 | 0.16 |
ENSRNOT00000037530
|
Mei4
|
meiotic double-stranded break formation protein 4 |
| chr7_+_41114697 | 0.15 |
ENSRNOT00000041354
|
Atp2b1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr2_-_202816562 | 0.14 |
ENSRNOT00000020401
|
Fam46c
|
family with sequence similarity 46, member C |
| chr5_+_147476221 | 0.14 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr2_-_106703401 | 0.14 |
ENSRNOT00000043933
|
AABR07009638.1
|
|
| chr7_+_41115154 | 0.14 |
ENSRNOT00000066174
|
Atp2b1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr11_+_33845463 | 0.13 |
ENSRNOT00000041838
|
Cbr1
|
carbonyl reductase 1 |
| chr7_-_144880092 | 0.13 |
ENSRNOT00000055281
|
Nfe2
|
nuclear factor, erythroid 2 |
| chr10_+_70884531 | 0.13 |
ENSRNOT00000015199
|
Ccl4
|
C-C motif chemokine ligand 4 |
| chr4_+_123586231 | 0.12 |
ENSRNOT00000049657
ENSRNOT00000013232 |
Grip2
|
glutamate receptor interacting protein 2 |
| chr1_+_213595240 | 0.12 |
ENSRNOT00000017137
|
Scgb1c1
|
secretoglobin, family 1C, member 1 |
| chr5_-_57896475 | 0.12 |
ENSRNOT00000017903
|
RGD1561916
|
similar to testes development-related NYD-SP22 isoform 1 |
| chr4_-_27966398 | 0.12 |
ENSRNOT00000012597
|
Cdk6
|
cyclin-dependent kinase 6 |
| chr11_+_33812989 | 0.12 |
ENSRNOT00000042283
ENSRNOT00000075985 |
Cbr1
|
carbonyl reductase 1 |
| chr1_+_141550633 | 0.11 |
ENSRNOT00000020047
|
Mesp2
|
mesoderm posterior bHLH transcription factor 2 |
| chr11_+_31389514 | 0.11 |
ENSRNOT00000000325
|
Olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr9_-_81566642 | 0.11 |
ENSRNOT00000080345
|
Aamp
|
angio-associated, migratory cell protein |
| chr1_-_169513537 | 0.10 |
ENSRNOT00000078058
|
Trim30c
|
tripartite motif-containing 30C |
| chr3_+_147226004 | 0.10 |
ENSRNOT00000012765
|
Tmem74b
|
transmembrane protein 74B |
| chr2_-_123851854 | 0.10 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr10_+_75032365 | 0.10 |
ENSRNOT00000010448
|
Supt4h1
|
SPT4 homolog, DSIF elongation factor subunit |
| chr5_+_144031402 | 0.10 |
ENSRNOT00000011694
|
Csf3r
|
colony stimulating factor 3 receptor |
| chr11_+_33909439 | 0.10 |
ENSRNOT00000002310
|
Cbr3
|
carbonyl reductase 3 |
| chrX_-_142248369 | 0.10 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr16_+_63837216 | 0.10 |
ENSRNOT00000014268
ENSRNOT00000014147 ENSRNOT00000038549 |
Nrg1
|
neuregulin 1 |
| chr1_+_141218095 | 0.10 |
ENSRNOT00000051411
|
LOC691427
|
similar to 6.8 kDa mitochondrial proteolipid |
| chr5_+_63192298 | 0.10 |
ENSRNOT00000008329
|
Sec61b
|
Sec61 translocon beta subunit |
| chr3_-_161131812 | 0.10 |
ENSRNOT00000067008
|
Wfdc11
|
WAP four-disulfide core domain 11 |
| chr8_+_106449321 | 0.10 |
ENSRNOT00000018622
|
Rbp1
|
retinol binding protein 1 |
| chr1_+_30681681 | 0.09 |
ENSRNOT00000015395
|
Rspo3
|
R-spondin 3 |
| chr10_-_18506337 | 0.09 |
ENSRNOT00000043036
|
Gabrp
|
gamma-aminobutyric acid type A receptor pi subunit |
| chr10_-_91661558 | 0.09 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
| chr3_-_102151489 | 0.09 |
ENSRNOT00000006349
|
Ano3
|
anoctamin 3 |
| chr1_-_247978540 | 0.09 |
ENSRNOT00000039428
|
RGD1311595
|
similar to KIAA2026 protein |
| chr11_+_33863500 | 0.09 |
ENSRNOT00000072384
|
LOC102556347
|
carbonyl reductase [NADPH] 1-like |
| chr10_-_70735742 | 0.09 |
ENSRNOT00000077035
|
Heatr9
|
HEAT repeat containing 9 |
| chr2_-_189856090 | 0.09 |
ENSRNOT00000020307
|
Npr1
|
natriuretic peptide receptor 1 |
| chr8_-_48652064 | 0.09 |
ENSRNOT00000030745
ENSRNOT00000083490 |
C2cd2l
|
C2CD2-like |
| chr1_+_85485289 | 0.09 |
ENSRNOT00000026182
|
Dll3
|
delta like canonical Notch ligand 3 |
| chr7_+_27240876 | 0.09 |
ENSRNOT00000091725
|
LOC362863
|
first gene upstream of Nt5dc3 |
| chr1_+_170242846 | 0.09 |
ENSRNOT00000023751
|
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr1_-_20155960 | 0.08 |
ENSRNOT00000061389
|
Samd3
|
sterile alpha motif domain containing 3 |
| chr13_-_83680207 | 0.08 |
ENSRNOT00000004199
|
Dcaf6
|
DDB1 and CUL4 associated factor 6 |
| chrX_-_17252877 | 0.08 |
ENSRNOT00000060213
|
LOC102547118
|
retinitis pigmentosa 1-like 1 protein-like |
| chrX_-_31759161 | 0.08 |
ENSRNOT00000041515
|
Asb9
|
ankyrin repeat and SOCS box-containing 9 |
| chr1_-_219438779 | 0.08 |
ENSRNOT00000029237
|
Tbc1d10c
|
TBC1 domain family, member 10C |
| chr4_+_21317695 | 0.08 |
ENSRNOT00000007572
|
Grm3
|
glutamate metabotropic receptor 3 |
| chr1_-_101236065 | 0.08 |
ENSRNOT00000066834
|
Cd37
|
CD37 molecule |
| chr7_+_94130852 | 0.08 |
ENSRNOT00000011485
|
Mal2
|
mal, T-cell differentiation protein 2 |
| chr4_+_121046277 | 0.08 |
ENSRNOT00000039058
|
Prr20e
|
proline rich 20E |
| chr1_-_260638816 | 0.08 |
ENSRNOT00000065632
ENSRNOT00000017938 ENSRNOT00000077962 |
Pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr7_+_145117951 | 0.08 |
ENSRNOT00000055272
|
Pde1b
|
phosphodiesterase 1B |
| chr8_-_22785671 | 0.07 |
ENSRNOT00000045384
|
Spc24
|
SPC24, NDC80 kinetochore complex component |
| chr10_+_56445647 | 0.07 |
ENSRNOT00000056870
|
Tmem256
|
transmembrane protein 256 |
| chr14_-_92495894 | 0.07 |
ENSRNOT00000064483
|
Cobl
|
cordon-bleu WH2 repeat protein |
| chr4_-_10517832 | 0.07 |
ENSRNOT00000039953
ENSRNOT00000083964 |
Gsap
|
gamma-secretase activating protein |
| chr4_+_28972434 | 0.07 |
ENSRNOT00000014219
|
Gngt1
|
G protein subunit gamma transducin 1 |
| chr3_-_39596718 | 0.07 |
ENSRNOT00000006784
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr1_-_116567189 | 0.07 |
ENSRNOT00000048996
|
LOC100912195
|
protein BEX1-like |
| chr1_-_261446570 | 0.07 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr3_+_93920447 | 0.07 |
ENSRNOT00000012625
|
Lmo2
|
LIM domain only 2 |
| chr5_+_74786480 | 0.07 |
ENSRNOT00000076889
|
Rn50_5_0791.1
|
|
| chr1_-_81946714 | 0.07 |
ENSRNOT00000027578
|
Grik5
|
glutamate ionotropic receptor kainate type subunit 5 |
| chr11_+_33812662 | 0.07 |
ENSRNOT00000085961
|
LOC102556347
|
carbonyl reductase [NADPH] 1-like |
| chrX_+_33443186 | 0.07 |
ENSRNOT00000005622
|
S100g
|
S100 calcium binding protein G |
| chr10_+_109774878 | 0.07 |
ENSRNOT00000054954
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr8_-_64305330 | 0.07 |
ENSRNOT00000030199
|
Tmem202
|
transmembrane protein 202 |
| chr20_+_7330250 | 0.07 |
ENSRNOT00000090993
|
LOC499407
|
LRRGT00097 |
| chr1_-_222182721 | 0.06 |
ENSRNOT00000078008
|
Tex40
|
testis expressed 40 |
| chr5_-_160405050 | 0.06 |
ENSRNOT00000081899
|
Ctrc
|
chymotrypsin C |
| chr16_+_63880428 | 0.06 |
ENSRNOT00000058727
|
Nrg1
|
neuregulin 1 |
| chr1_-_79930263 | 0.06 |
ENSRNOT00000019219
|
Foxa3
|
forkhead box A3 |
| chr8_+_94693290 | 0.06 |
ENSRNOT00000013908
|
Cyb5r4
|
cytochrome b5 reductase 4 |
| chr13_-_85443727 | 0.06 |
ENSRNOT00000090668
ENSRNOT00000076125 |
Uck2
|
uridine-cytidine kinase 2 |
| chr6_-_2311781 | 0.06 |
ENSRNOT00000084171
|
Cyp1b1
|
cytochrome P450, family 1, subfamily b, polypeptide 1 |
| chr9_+_77320726 | 0.06 |
ENSRNOT00000068450
|
Spag16
|
sperm associated antigen 16 |
| chr20_-_12858096 | 0.06 |
ENSRNOT00000085270
|
Lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr1_+_243589607 | 0.06 |
ENSRNOT00000021792
|
Dmrt3
|
doublesex and mab-3 related transcription factor 3 |
| chr12_-_38516475 | 0.06 |
ENSRNOT00000082462
|
Lrrc43
|
leucine rich repeat containing 43 |
| chr17_-_10559627 | 0.06 |
ENSRNOT00000023515
|
Higd2a
|
HIG1 hypoxia inducible domain family, member 2A |
| chr9_+_81868265 | 0.06 |
ENSRNOT00000022632
|
Bcs1l
|
BCS1 homolog, ubiquinol-cytochrome c reductase complex chaperone |
| chr8_+_62779875 | 0.06 |
ENSRNOT00000010831
|
Cyp11a1
|
cytochrome P450, family 11, subfamily a, polypeptide 1 |
| chr10_+_69434941 | 0.06 |
ENSRNOT00000009756
|
Ccl11
|
C-C motif chemokine ligand 11 |
| chr7_-_29949603 | 0.06 |
ENSRNOT00000009419
|
LOC103692829
|
60S ribosomal protein L9 pseudogene |
| chr19_-_55284663 | 0.06 |
ENSRNOT00000018387
|
Snai3
|
snail family transcriptional repressor 3 |
| chr5_+_173248245 | 0.06 |
ENSRNOT00000025213
|
Mrpl20
|
mitochondrial ribosomal protein L20 |
| chr18_+_16616937 | 0.06 |
ENSRNOT00000093641
|
Mocos
|
molybdenum cofactor sulfurase |
| chr15_-_4399589 | 0.06 |
ENSRNOT00000065858
ENSRNOT00000079812 |
Fam149b1
|
family with sequence similarity 149, member B1 |
| chr3_+_4282236 | 0.06 |
ENSRNOT00000042201
|
LOC100364457
|
ribosomal protein L9-like |
| chr7_+_77763512 | 0.05 |
ENSRNOT00000006411
|
Baalc
|
brain and acute leukemia, cytoplasmic |
| chr1_-_53087474 | 0.05 |
ENSRNOT00000017302
|
Ccr6
|
C-C motif chemokine receptor 6 |
| chr13_+_52662996 | 0.05 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr8_-_52814188 | 0.05 |
ENSRNOT00000087265
ENSRNOT00000047633 |
Rexo2
|
RNA exonuclease 2 |
| chr19_+_24496569 | 0.05 |
ENSRNOT00000004982
|
Mgat4d
|
MGAT4 family, member D |
| chr7_-_12905339 | 0.05 |
ENSRNOT00000088330
|
Cdc34
|
cell division cycle 34 |
| chr17_+_10559680 | 0.05 |
ENSRNOT00000023320
|
Nop16
|
NOP16 nucleolar protein |
| chr7_-_3246071 | 0.05 |
ENSRNOT00000044292
|
Ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr10_-_41408518 | 0.05 |
ENSRNOT00000018967
|
Nmur2
|
neuromedin U receptor 2 |
| chr2_+_198655437 | 0.05 |
ENSRNOT00000028781
|
Hfe2
|
hemochromatosis type 2 (juvenile) |
| chr7_+_120580743 | 0.05 |
ENSRNOT00000017181
|
Maff
|
MAF bZIP transcription factor F |
| chr5_+_159671136 | 0.05 |
ENSRNOT00000084737
|
Fbxo42
|
F-box protein 42 |
| chr20_-_3401273 | 0.05 |
ENSRNOT00000089257
ENSRNOT00000078451 ENSRNOT00000001085 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr9_+_30419001 | 0.05 |
ENSRNOT00000018116
|
Col9a1
|
collagen type IX alpha 1 chain |
| chr8_-_110813000 | 0.05 |
ENSRNOT00000010634
|
Ephb1
|
Eph receptor B1 |
| chr4_-_14490446 | 0.05 |
ENSRNOT00000009132
|
Sema3c
|
semaphorin 3C |
| chr9_+_98505259 | 0.05 |
ENSRNOT00000051810
|
Erfe
|
erythroferrone |
| chr1_-_101803200 | 0.05 |
ENSRNOT00000028588
|
Cyth2
|
cytohesin 2 |
| chr6_+_109466060 | 0.05 |
ENSRNOT00000011339
|
Jdp2
|
Jun dimerization protein 2 |
| chr9_+_44491177 | 0.05 |
ENSRNOT00000075242
|
NEWGENE_1308196
|
mitochondrial ribosomal protein L30 |
| chr15_+_32894938 | 0.05 |
ENSRNOT00000012837
|
Abhd4
|
abhydrolase domain containing 4 |
| chr14_+_59611434 | 0.05 |
ENSRNOT00000065366
|
Cckar
|
cholecystokinin A receptor |
| chr5_-_173680290 | 0.05 |
ENSRNOT00000027564
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr8_-_84522588 | 0.05 |
ENSRNOT00000076213
|
Mlip
|
muscular LMNA-interacting protein |
| chr13_-_50509916 | 0.05 |
ENSRNOT00000076747
|
Ren
|
renin |
| chr2_-_23289266 | 0.05 |
ENSRNOT00000061708
|
Bhmt2
|
betaine-homocysteine S-methyltransferase 2 |
| chr1_+_53174879 | 0.05 |
ENSRNOT00000017601
ENSRNOT00000084628 |
Rnaset2
|
ribonuclease T2 |
| chr10_-_88152064 | 0.05 |
ENSRNOT00000019477
|
Krt16
|
keratin 16 |
| chrX_-_111102464 | 0.05 |
ENSRNOT00000084176
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr1_-_222098297 | 0.05 |
ENSRNOT00000028678
|
Rps6ka4
|
ribosomal protein S6 kinase A4 |
| chr1_-_153740905 | 0.05 |
ENSRNOT00000023239
|
Prss23
|
protease, serine, 23 |
| chr4_-_119568736 | 0.05 |
ENSRNOT00000041234
|
Aplf
|
aprataxin and PNKP like factor |
| chr1_+_142951094 | 0.05 |
ENSRNOT00000077441
|
Slc28a1
|
solute carrier family 28 member 1 |
| chr7_+_3246220 | 0.05 |
ENSRNOT00000009155
|
Sarnp
|
SAP domain containing ribonucleoprotein |
| chr9_+_100511610 | 0.05 |
ENSRNOT00000033233
|
Ano7
|
anoctamin 7 |
| chr1_+_185863043 | 0.05 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr9_+_82596355 | 0.05 |
ENSRNOT00000065076
|
Speg
|
SPEG complex locus |
| chr1_-_178195948 | 0.05 |
ENSRNOT00000081767
|
Btbd10
|
BTB domain containing 10 |
| chr7_+_14891001 | 0.04 |
ENSRNOT00000041034
|
Cyp4f4
|
cytochrome P450, family 4, subfamily f, polypeptide 4 |
| chr1_+_91042635 | 0.04 |
ENSRNOT00000028211
|
LOC103690005
|
tubulin-folding cofactor B |
| chr14_-_14390699 | 0.04 |
ENSRNOT00000046639
|
Anxa3
|
annexin A3 |
| chr1_-_178196294 | 0.04 |
ENSRNOT00000067170
|
Btbd10
|
BTB domain containing 10 |
| chr6_-_50923510 | 0.04 |
ENSRNOT00000010631
|
Bcap29
|
B-cell receptor-associated protein 29 |
| chr7_+_14529482 | 0.04 |
ENSRNOT00000090844
ENSRNOT00000060284 ENSRNOT00000093239 |
Cyp4f5
Cyp4f17
|
cytochrome P450, family 4, subfamily f, polypeptide 5 cytochrome P450, family 4, subfamily f, polypeptide 17 |
| chr16_+_14306804 | 0.04 |
ENSRNOT00000017773
|
Lrit1
|
leucine-rich repeat, Ig-like and transmembrane domains 1 |
| chr1_-_87155118 | 0.04 |
ENSRNOT00000072441
|
AABR07002854.1
|
|
| chr1_-_22404002 | 0.04 |
ENSRNOT00000044098
|
Taar8a
|
trace amine-associated receptor 8a |
| chr1_+_104106245 | 0.04 |
ENSRNOT00000019175
ENSRNOT00000081660 |
Zdhhc13
|
zinc finger, DHHC-type containing 13 |
| chr16_-_18645941 | 0.04 |
ENSRNOT00000079241
|
Dydc2
|
DPY30 domain containing 2 |
| chr15_+_2527343 | 0.04 |
ENSRNOT00000089013
|
Dusp13
|
dual specificity phosphatase 13 |
| chr20_-_45259928 | 0.04 |
ENSRNOT00000087226
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr11_+_27208564 | 0.04 |
ENSRNOT00000002158
|
Map3k7cl
|
MAP3K7 C-terminal like |
| chr1_-_141533908 | 0.04 |
ENSRNOT00000020117
|
Mesp1
|
mesoderm posterior bHLH transcription factor 1 |
| chr3_+_5624506 | 0.04 |
ENSRNOT00000036995
|
Adamtsl2
|
ADAMTS-like 2 |
| chr19_-_26129619 | 0.04 |
ENSRNOT00000067600
|
Best2
|
bestrophin 2 |
| chr18_-_69944632 | 0.04 |
ENSRNOT00000047271
|
Mapk4
|
mitogen-activated protein kinase 4 |
| chr15_+_2734068 | 0.04 |
ENSRNOT00000017663
|
Dusp13
|
dual specificity phosphatase 13 |
| chr2_-_59084059 | 0.04 |
ENSRNOT00000086323
ENSRNOT00000088701 |
Spef2
|
sperm flagellar 2 |
| chr18_-_29611212 | 0.04 |
ENSRNOT00000022685
|
Dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr1_+_85380088 | 0.04 |
ENSRNOT00000090129
|
Zfp36
|
zinc finger protein 36 |
| chr15_+_40937708 | 0.04 |
ENSRNOT00000018407
|
Spata13
|
spermatogenesis associated 13 |
| chr18_-_70184337 | 0.04 |
ENSRNOT00000020637
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr5_-_141430659 | 0.04 |
ENSRNOT00000034944
|
Akirin1
|
akirin 1 |
| chr15_-_24199341 | 0.04 |
ENSRNOT00000015553
|
Dlgap5
|
DLG associated protein 5 |
| chr10_-_50402616 | 0.04 |
ENSRNOT00000004546
|
Hs3st3b1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1 |
| chr3_-_151600140 | 0.04 |
ENSRNOT00000026451
|
Fer1l4
|
fer-1-like family member 4 |
| chr9_-_9675110 | 0.04 |
ENSRNOT00000073294
|
Vav1
|
vav guanine nucleotide exchange factor 1 |
| chr7_-_12646960 | 0.04 |
ENSRNOT00000014687
|
Prtn3
|
proteinase 3 |
| chr16_+_26906716 | 0.04 |
ENSRNOT00000064297
|
Cpe
|
carboxypeptidase E |
| chr4_+_7257752 | 0.04 |
ENSRNOT00000088357
|
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr1_+_196996581 | 0.04 |
ENSRNOT00000021690
|
Il21r
|
interleukin 21 receptor |
| chr16_+_81153489 | 0.04 |
ENSRNOT00000024999
|
Grk1
|
G protein-coupled receptor kinase 1 |
| chr18_+_29639872 | 0.04 |
ENSRNOT00000021516
|
Zmat2
|
zinc finger, matrin type 2 |
| chr3_-_160891190 | 0.04 |
ENSRNOT00000019386
|
Sdc4
|
syndecan 4 |
| chr6_-_6842758 | 0.04 |
ENSRNOT00000006094
|
Kcng3
|
potassium voltage-gated channel modifier subfamily G member 3 |
| chr19_+_24545318 | 0.04 |
ENSRNOT00000005071
|
Clgn
|
calmegin |
| chr13_-_85443976 | 0.04 |
ENSRNOT00000005213
|
Uck2
|
uridine-cytidine kinase 2 |
| chr1_+_277689729 | 0.04 |
ENSRNOT00000051834
|
Vwa2
|
von Willebrand factor A domain containing 2 |
| chr13_+_71656651 | 0.04 |
ENSRNOT00000050365
|
Zfp648
|
zinc finger protein 648 |
| chr15_-_12129220 | 0.04 |
ENSRNOT00000029318
|
Olr1605
|
olfactory receptor 1605 |
| chr5_+_136014017 | 0.04 |
ENSRNOT00000025346
|
Best4
|
bestrophin 4 |
| chr20_+_44436403 | 0.04 |
ENSRNOT00000000733
ENSRNOT00000076859 |
Fyn
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr5_+_157327657 | 0.04 |
ENSRNOT00000022847
|
Pla2g2e
|
phospholipase A2, group IIE |
| chr7_-_27240528 | 0.04 |
ENSRNOT00000029435
ENSRNOT00000079731 |
Hsp90b1
|
heat shock protein 90 beta family member 1 |
| chr9_+_95241609 | 0.03 |
ENSRNOT00000032634
ENSRNOT00000077416 |
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr3_-_59920935 | 0.03 |
ENSRNOT00000026040
|
Ola1
|
Obg-like ATPase 1 |
| chr3_-_113423149 | 0.03 |
ENSRNOT00000021008
|
Serinc4
|
serine incorporator 4 |
| chr4_-_176026133 | 0.03 |
ENSRNOT00000043374
ENSRNOT00000046598 |
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr17_-_86322144 | 0.03 |
ENSRNOT00000034474
|
LOC681241
|
hypothetical protein LOC681241 |
| chr12_+_40466495 | 0.03 |
ENSRNOT00000001816
|
Aldh2
|
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr19_+_39063998 | 0.03 |
ENSRNOT00000081116
|
Has3
|
hyaluronan synthase 3 |
| chr5_-_139864393 | 0.03 |
ENSRNOT00000030016
|
Zfp69
|
zinc finger protein 69 |
| chr2_-_197982385 | 0.03 |
ENSRNOT00000034074
|
Mrps21
|
mitochondrial ribosomal protein S21 |
| chr6_+_37144787 | 0.03 |
ENSRNOT00000075503
|
Rad51ap2
|
RAD51 associated protein 2 |
| chrX_-_153491837 | 0.03 |
ENSRNOT00000089027
|
LOC100360413
|
eukaryotic translation elongation factor 1 alpha 1-like |
| chr1_-_206394346 | 0.03 |
ENSRNOT00000038122
|
LOC100302465
|
hypothetical LOC100302465 |
| chr2_+_93520734 | 0.03 |
ENSRNOT00000083909
|
Snx16
|
sorting nexin 16 |
| chr9_+_81689802 | 0.03 |
ENSRNOT00000021432
|
Vil1
|
villin 1 |
| chr7_-_2786856 | 0.03 |
ENSRNOT00000047530
ENSRNOT00000086939 |
Coq10a
|
coenzyme Q10A |
| chr20_+_13498926 | 0.03 |
ENSRNOT00000070992
ENSRNOT00000045375 |
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr5_+_166870011 | 0.03 |
ENSRNOT00000088299
|
Rps2
|
ribosomal protein S2 |
| chrX_-_154918095 | 0.03 |
ENSRNOT00000085224
|
LOC100911991
|
elongation factor 1-alpha 1-like |
| chr4_-_82263302 | 0.03 |
ENSRNOT00000082933
|
LOC100909604
|
homeobox protein Hox-A6-like |
| chr2_-_80408672 | 0.03 |
ENSRNOT00000067638
|
Fam105a
|
family with sequence similarity 105, member A |
| chr4_+_147275334 | 0.03 |
ENSRNOT00000051858
|
Pparg
|
peroxisome proliferator-activated receptor gamma |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.2 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.1 | 0.3 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.2 | GO:2000853 | negative regulation of corticosterone secretion(GO:2000853) |
| 0.0 | 0.2 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.0 | 0.3 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 0.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:2000040 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0071387 | trabecular meshwork development(GO:0002930) cellular response to cortisol stimulus(GO:0071387) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.0 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.0 | GO:0060060 | photoreceptor cell morphogenesis(GO:0008594) post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.0 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.0 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.0 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.0 | GO:1904823 | purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.0 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.0 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.0 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0031559 | lanosterol synthase activity(GO:0000250) oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |