Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
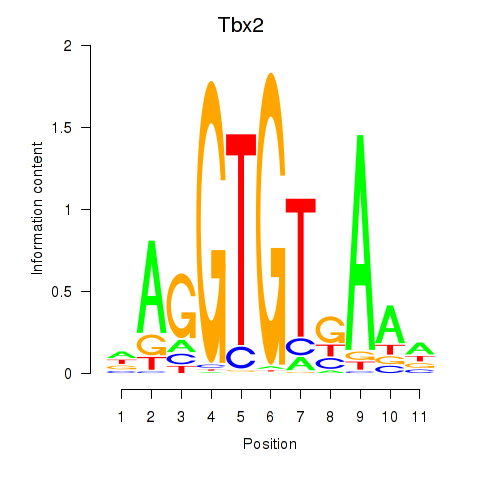
Results for Tbx2
Z-value: 0.52
Transcription factors associated with Tbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx2
|
ENSRNOG00000003517 | T-box 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx2 | rn6_v1_chr10_+_73279119_73279119 | -0.99 | 1.8e-03 | Click! |
Activity profile of Tbx2 motif
Sorted Z-values of Tbx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_141550633 | 0.35 |
ENSRNOT00000020047
|
Mesp2
|
mesoderm posterior bHLH transcription factor 2 |
| chr1_-_20155960 | 0.32 |
ENSRNOT00000061389
|
Samd3
|
sterile alpha motif domain containing 3 |
| chr5_+_50381244 | 0.28 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr8_+_52829085 | 0.25 |
ENSRNOT00000007754
|
RGD1563941
|
similar to hypothetical protein FLJ20010 |
| chr20_+_6356423 | 0.24 |
ENSRNOT00000000628
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr2_-_45077219 | 0.21 |
ENSRNOT00000014319
|
Gzmk
|
granzyme K |
| chr13_-_55878094 | 0.20 |
ENSRNOT00000014218
|
Lhx9
|
LIM homeobox 9 |
| chr4_-_148845267 | 0.17 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chrX_-_111102464 | 0.15 |
ENSRNOT00000084176
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr20_-_4677506 | 0.15 |
ENSRNOT00000061297
|
RT1-CE6
|
RT1 class I, locus CE6 |
| chr11_-_61530567 | 0.14 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr14_-_21709084 | 0.14 |
ENSRNOT00000087477
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chr8_-_75995371 | 0.13 |
ENSRNOT00000014014
|
Foxb1
|
forkhead box B1 |
| chr1_+_85485289 | 0.13 |
ENSRNOT00000026182
|
Dll3
|
delta like canonical Notch ligand 3 |
| chr1_-_124803363 | 0.12 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr16_-_49574314 | 0.11 |
ENSRNOT00000017568
ENSRNOT00000085535 ENSRNOT00000017054 |
Pdlim3
|
PDZ and LIM domain 3 |
| chr9_+_98505259 | 0.10 |
ENSRNOT00000051810
|
Erfe
|
erythroferrone |
| chr10_+_49541051 | 0.10 |
ENSRNOT00000041606
|
Pmp22
|
peripheral myelin protein 22 |
| chr3_-_164388492 | 0.09 |
ENSRNOT00000090493
|
Tmem189
|
transmembrane protein 189 |
| chr2_-_210738378 | 0.09 |
ENSRNOT00000025746
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr14_-_14390699 | 0.09 |
ENSRNOT00000046639
|
Anxa3
|
annexin A3 |
| chr7_-_120839577 | 0.09 |
ENSRNOT00000018526
|
Dmc1
|
DNA meiotic recombinase 1 |
| chr1_-_260341723 | 0.08 |
ENSRNOT00000030656
|
Opalin
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr12_+_40466495 | 0.08 |
ENSRNOT00000001816
|
Aldh2
|
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr1_+_89017479 | 0.08 |
ENSRNOT00000038154
|
U2af1l4
|
U2 small nuclear RNA auxiliary factor 1-like 4 |
| chr1_+_185863043 | 0.08 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr18_+_16616937 | 0.08 |
ENSRNOT00000093641
|
Mocos
|
molybdenum cofactor sulfurase |
| chr5_+_24434872 | 0.08 |
ENSRNOT00000010696
|
Ccne2
|
cyclin E2 |
| chr1_-_89017290 | 0.08 |
ENSRNOT00000028438
|
Psenen
|
presenilin enhancer gamma secretase subunit |
| chr1_+_21525421 | 0.08 |
ENSRNOT00000017911
|
Arg1
|
arginase 1 |
| chr8_+_58431407 | 0.07 |
ENSRNOT00000011974
|
Sln
|
sarcolipin |
| chr2_+_198755262 | 0.07 |
ENSRNOT00000028807
|
Rbm8a
|
RNA binding motif protein 8A |
| chr1_-_141533908 | 0.07 |
ENSRNOT00000020117
|
Mesp1
|
mesoderm posterior bHLH transcription factor 1 |
| chr1_+_220243473 | 0.07 |
ENSRNOT00000084650
ENSRNOT00000027091 |
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr2_-_251532312 | 0.07 |
ENSRNOT00000019501
|
Cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr4_-_145678066 | 0.07 |
ENSRNOT00000014103
|
Ghrl
|
ghrelin and obestatin prepropeptide |
| chr3_+_170994038 | 0.07 |
ENSRNOT00000081823
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chr10_+_63884338 | 0.07 |
ENSRNOT00000007100
|
Ywhae
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon |
| chr3_+_122720769 | 0.06 |
ENSRNOT00000009323
|
Tmc2
|
transmembrane channel-like 2 |
| chrX_+_128897181 | 0.06 |
ENSRNOT00000008444
|
Sh2d1a
|
SH2 domain containing 1A |
| chr19_-_25288335 | 0.06 |
ENSRNOT00000050214
|
Nanos3
|
nanos C2HC-type zinc finger 3 |
| chrX_+_70563570 | 0.05 |
ENSRNOT00000003772
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chrX_+_135348436 | 0.05 |
ENSRNOT00000008868
|
Rab33a
|
RAB33A, member RAS oncogene family |
| chr20_-_4677034 | 0.05 |
ENSRNOT00000075244
|
Rn60_20_0047.3
|
|
| chr1_+_22332090 | 0.05 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chr4_+_113887115 | 0.05 |
ENSRNOT00000010602
|
Aup1
|
ancient ubiquitous protein 1 |
| chr5_+_57845819 | 0.05 |
ENSRNOT00000017712
|
Nudt2
|
nudix hydrolase 2 |
| chr1_-_89559960 | 0.05 |
ENSRNOT00000092133
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr5_+_164808323 | 0.05 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr7_-_12635943 | 0.05 |
ENSRNOT00000015029
|
Cfd
|
complement factor D |
| chr14_-_84662143 | 0.05 |
ENSRNOT00000057529
ENSRNOT00000080078 |
Hormad2
|
HORMA domain containing 2 |
| chr20_-_3728844 | 0.05 |
ENSRNOT00000074958
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 |
| chr2_+_51672722 | 0.05 |
ENSRNOT00000016485
|
Fgf10
|
fibroblast growth factor 10 |
| chr18_+_53609214 | 0.05 |
ENSRNOT00000077443
|
Slc27a6
|
solute carrier family 27 member 6 |
| chr19_+_12097632 | 0.05 |
ENSRNOT00000049452
|
Ces5a
|
carboxylesterase 5A |
| chr4_-_114853868 | 0.05 |
ENSRNOT00000090828
|
Wdr54
|
WD repeat domain 54 |
| chr17_-_14828329 | 0.04 |
ENSRNOT00000022830
|
Barx1
|
BARX homeobox 1 |
| chr13_-_95943761 | 0.04 |
ENSRNOT00000005961
|
Adss
|
adenylosuccinate synthase |
| chr15_+_28295368 | 0.04 |
ENSRNOT00000013786
|
Slc39a2
|
solute carrier family 39 member 2 |
| chr13_-_85443976 | 0.04 |
ENSRNOT00000005213
|
Uck2
|
uridine-cytidine kinase 2 |
| chr9_+_47225585 | 0.04 |
ENSRNOT00000077303
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr1_-_22404002 | 0.04 |
ENSRNOT00000044098
|
Taar8a
|
trace amine-associated receptor 8a |
| chr4_-_113988246 | 0.04 |
ENSRNOT00000013128
|
LOC103692166
|
WD repeat-containing protein 54 |
| chr13_+_82429063 | 0.04 |
ENSRNOT00000076879
|
Selp
|
selectin P |
| chr11_+_87677966 | 0.04 |
ENSRNOT00000068711
|
Klhl22
|
kelch-like family member 22 |
| chr6_+_109300433 | 0.04 |
ENSRNOT00000010712
|
Fos
|
FBJ osteosarcoma oncogene |
| chr6_-_103605256 | 0.03 |
ENSRNOT00000006005
|
Dcaf5
|
DDB1 and CUL4 associated factor 5 |
| chr1_+_277689729 | 0.03 |
ENSRNOT00000051834
|
Vwa2
|
von Willebrand factor A domain containing 2 |
| chr7_-_122926336 | 0.03 |
ENSRNOT00000000205
|
Chadl
|
chondroadherin-like |
| chr6_-_44363915 | 0.03 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr8_+_42511520 | 0.03 |
ENSRNOT00000042974
|
Olr1259
|
olfactory receptor 1259 |
| chr13_+_49092306 | 0.03 |
ENSRNOT00000000039
|
Nuak2
|
NUAK family kinase 2 |
| chr12_-_3924415 | 0.03 |
ENSRNOT00000067752
|
AABR07035074.1
|
|
| chr5_-_150704117 | 0.03 |
ENSRNOT00000067455
|
Sesn2
|
sestrin 2 |
| chrX_+_104734082 | 0.03 |
ENSRNOT00000005020
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr5_-_74029238 | 0.03 |
ENSRNOT00000031432
|
Frrs1l
|
ferric-chelate reductase 1-like |
| chr1_+_22364551 | 0.03 |
ENSRNOT00000043157
|
LOC108348200
|
trace amine-associated receptor 8a |
| chr9_+_70310932 | 0.02 |
ENSRNOT00000088268
|
LOC103690541
|
uncharacterized LOC103690541 |
| chr19_+_24496569 | 0.02 |
ENSRNOT00000004982
|
Mgat4d
|
MGAT4 family, member D |
| chrX_-_72370044 | 0.02 |
ENSRNOT00000004224
|
Hdac8
|
histone deacetylase 8 |
| chr1_+_213686046 | 0.02 |
ENSRNOT00000019808
|
LOC108348167
|
NACHT, LRR and PYD domains-containing protein 6-like |
| chr13_-_85443727 | 0.02 |
ENSRNOT00000090668
ENSRNOT00000076125 |
Uck2
|
uridine-cytidine kinase 2 |
| chr8_-_13680870 | 0.02 |
ENSRNOT00000081616
|
Hephl1
|
hephaestin-like 1 |
| chr1_+_213676954 | 0.02 |
ENSRNOT00000050551
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr17_-_89163113 | 0.02 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr4_+_114854458 | 0.02 |
ENSRNOT00000013312
|
LOC500227
|
hypothetical gene supported by BC079424 |
| chr16_+_61130755 | 0.02 |
ENSRNOT00000042609
|
AABR07026048.1
|
|
| chr19_-_31867850 | 0.02 |
ENSRNOT00000024689
|
Anapc10
|
anaphase promoting complex subunit 10 |
| chr4_-_70973842 | 0.02 |
ENSRNOT00000051687
|
Trpv5
|
transient receptor potential cation channel, subfamily V, member 5 |
| chr10_-_88645364 | 0.01 |
ENSRNOT00000030344
|
Rab5c
|
RAB5C, member RAS oncogene family |
| chr15_-_34322115 | 0.01 |
ENSRNOT00000026734
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr5_-_171415354 | 0.01 |
ENSRNOT00000055398
|
Tp73
|
tumor protein p73 |
| chrX_+_33599671 | 0.01 |
ENSRNOT00000006843
|
Txlng
|
taxilin gamma |
| chr15_-_51386190 | 0.01 |
ENSRNOT00000022706
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chrX_-_142248369 | 0.01 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr1_+_100224401 | 0.01 |
ENSRNOT00000025846
|
Fv1
|
Friend virus susceptibility 1 |
| chr1_-_226353611 | 0.01 |
ENSRNOT00000037624
|
Dagla
|
diacylglycerol lipase, alpha |
| chr17_+_72240802 | 0.01 |
ENSRNOT00000025993
ENSRNOT00000090453 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr7_+_143679617 | 0.01 |
ENSRNOT00000014033
|
Eif4b
|
eukaryotic translation initiation factor 4B |
| chr9_+_61738471 | 0.00 |
ENSRNOT00000090305
|
AABR07067762.1
|
|
| chr11_+_75905443 | 0.00 |
ENSRNOT00000002650
|
Fgf12
|
fibroblast growth factor 12 |
| chrX_+_68713123 | 0.00 |
ENSRNOT00000076690
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr1_+_238843338 | 0.00 |
ENSRNOT00000024583
|
Zfand5
|
zinc finger AN1-type containing 5 |
| chrX_+_134979646 | 0.00 |
ENSRNOT00000006035
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr5_+_113592919 | 0.00 |
ENSRNOT00000011336
|
Ift74
|
intraflagellar transport 74 |
| chr7_-_141624972 | 0.00 |
ENSRNOT00000078785
|
AABR07058884.1
|
|
| chr1_+_85003280 | 0.00 |
ENSRNOT00000057122
|
Fcgbp
|
Fc fragment of IgG binding protein |
| chr1_+_80777014 | 0.00 |
ENSRNOT00000079758
|
Gm19345
|
predicted gene, 19345 |
| chr19_+_33932532 | 0.00 |
ENSRNOT00000078395
ENSRNOT00000091985 ENSRNOT00000078701 ENSRNOT00000080317 |
Ednra
|
endothelin receptor type A |
| chr7_-_11406771 | 0.00 |
ENSRNOT00000047450
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr4_-_52350624 | 0.00 |
ENSRNOT00000060476
|
Tmem229a
|
transmembrane protein 229A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.2 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.2 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.3 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.1 | GO:0051464 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) negative regulation of interleukin-6 biosynthetic process(GO:0045409) positive regulation of cortisol secretion(GO:0051464) |
| 0.0 | 0.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:1902023 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.0 | 0.0 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0051177 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.0 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.1 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 0.0 | 0.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |