Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Tbx1_Eomes
Z-value: 0.34
Transcription factors associated with Tbx1_Eomes
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
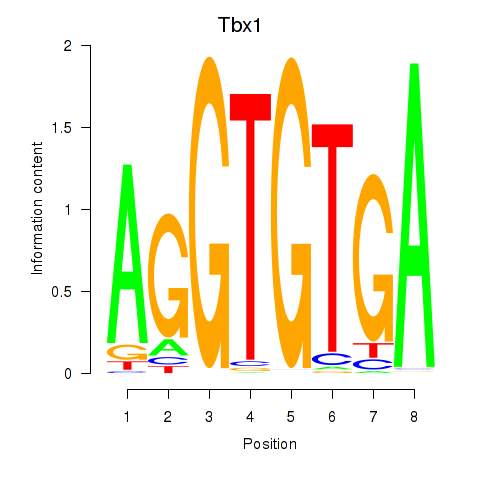
Tbx1
|
ENSRNOG00000001892 | T-box 1 |
|
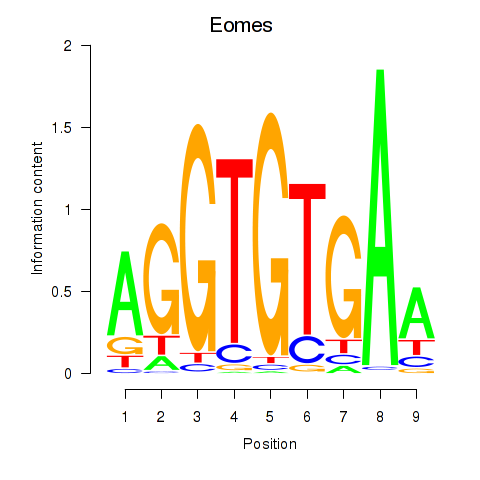
Eomes
|
ENSRNOG00000042140 | eomesodermin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Eomes | rn6_v1_chr8_+_127144903_127144903 | 0.89 | 4.6e-02 | Click! |
| Tbx1 | rn6_v1_chr11_+_86552022_86552022 | 0.76 | 1.3e-01 | Click! |
Activity profile of Tbx1_Eomes motif
Sorted Z-values of Tbx1_Eomes motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_227047290 | 0.12 |
ENSRNOT00000037226
|
Ms4a15
|
membrane spanning 4-domains A15 |
| chr6_+_28323647 | 0.10 |
ENSRNOT00000089093
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr1_-_124803363 | 0.10 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr9_-_63637677 | 0.09 |
ENSRNOT00000049259
|
Satb2
|
SATB homeobox 2 |
| chr18_+_53609214 | 0.09 |
ENSRNOT00000077443
|
Slc27a6
|
solute carrier family 27 member 6 |
| chr12_+_52359310 | 0.08 |
ENSRNOT00000071857
ENSRNOT00000065893 |
Fbrsl1
|
fibrosin-like 1 |
| chr16_+_46731403 | 0.08 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr4_-_180234804 | 0.08 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr10_-_88754829 | 0.07 |
ENSRNOT00000026354
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr3_+_161433410 | 0.07 |
ENSRNOT00000024657
|
Slc12a5
|
solute carrier family 12 member 5 |
| chr9_-_15700235 | 0.07 |
ENSRNOT00000088713
ENSRNOT00000035907 |
Trerf1
|
transcriptional regulating factor 1 |
| chr10_-_88645364 | 0.07 |
ENSRNOT00000030344
|
Rab5c
|
RAB5C, member RAS oncogene family |
| chr19_-_25288335 | 0.07 |
ENSRNOT00000050214
|
Nanos3
|
nanos C2HC-type zinc finger 3 |
| chr5_+_136683592 | 0.06 |
ENSRNOT00000085527
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr3_+_131351587 | 0.06 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr2_+_207262934 | 0.06 |
ENSRNOT00000016833
|
Ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr8_-_45215974 | 0.06 |
ENSRNOT00000010792
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr5_+_143500441 | 0.05 |
ENSRNOT00000045513
|
Grik3
|
glutamate ionotropic receptor kainate type subunit 3 |
| chr20_+_6351458 | 0.05 |
ENSRNOT00000091731
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr6_-_44363915 | 0.05 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr17_-_14091766 | 0.04 |
ENSRNOT00000075373
|
AABR07027088.1
|
|
| chr1_-_20155960 | 0.04 |
ENSRNOT00000061389
|
Samd3
|
sterile alpha motif domain containing 3 |
| chr1_+_187149453 | 0.04 |
ENSRNOT00000082738
|
Xylt1
|
xylosyltransferase 1 |
| chr8_-_78233430 | 0.04 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr2_-_177924970 | 0.04 |
ENSRNOT00000029340
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr4_+_146455332 | 0.04 |
ENSRNOT00000009775
|
Hrh1
|
histamine receptor H 1 |
| chr1_+_238222521 | 0.04 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr15_+_8739589 | 0.04 |
ENSRNOT00000071751
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr5_+_63781801 | 0.03 |
ENSRNOT00000008302
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr5_-_12563429 | 0.03 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr1_+_79982639 | 0.03 |
ENSRNOT00000071916
|
Dmwd
|
dystrophia myotonica, WD repeat containing |
| chr18_-_28871849 | 0.03 |
ENSRNOT00000025902
|
Nrg2
|
neuregulin 2 |
| chr7_+_41475163 | 0.03 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr3_+_160231914 | 0.03 |
ENSRNOT00000014411
|
Kcnk15
|
potassium two pore domain channel subfamily K member 15 |
| chr2_+_263212051 | 0.03 |
ENSRNOT00000089396
|
Negr1
|
neuronal growth regulator 1 |
| chr17_-_80320681 | 0.03 |
ENSRNOT00000023637
|
C1ql3
|
complement C1q like 3 |
| chr5_-_150704117 | 0.03 |
ENSRNOT00000067455
|
Sesn2
|
sestrin 2 |
| chr6_+_101532518 | 0.03 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr8_+_32188617 | 0.03 |
ENSRNOT00000081145
|
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr5_+_58393603 | 0.02 |
ENSRNOT00000080082
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr11_+_47061354 | 0.02 |
ENSRNOT00000039997
|
Pcnp
|
PEST proteolytic signal containing nuclear protein |
| chr14_+_12218553 | 0.02 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr2_-_62634785 | 0.02 |
ENSRNOT00000017937
|
Pdzd2
|
PDZ domain containing 2 |
| chr7_-_27240528 | 0.02 |
ENSRNOT00000029435
ENSRNOT00000079731 |
Hsp90b1
|
heat shock protein 90 beta family member 1 |
| chr20_+_2501252 | 0.02 |
ENSRNOT00000079307
ENSRNOT00000084559 |
Trim39
|
tripartite motif-containing 39 |
| chr9_+_3896337 | 0.02 |
ENSRNOT00000079166
|
LOC103693189
|
protein tyrosine phosphatase type IVA 1 |
| chr11_-_4397361 | 0.02 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr11_+_61662270 | 0.02 |
ENSRNOT00000079521
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr1_-_264975132 | 0.02 |
ENSRNOT00000021748
|
Lbx1
|
ladybird homeobox 1 |
| chr4_-_100783016 | 0.02 |
ENSRNOT00000020671
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chrX_+_70563570 | 0.02 |
ENSRNOT00000003772
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr5_-_79874671 | 0.02 |
ENSRNOT00000084563
|
Tnc
|
tenascin C |
| chr18_+_46148849 | 0.02 |
ENSRNOT00000026724
|
Prr16
|
proline rich 16 |
| chr11_+_80255790 | 0.02 |
ENSRNOT00000002522
|
Bcl6
|
B-cell CLL/lymphoma 6 |
| chr1_-_72468738 | 0.02 |
ENSRNOT00000029136
|
Zfp628
|
zinc finger protein 628 |
| chr2_+_74360622 | 0.02 |
ENSRNOT00000014013
|
Cdh18
|
cadherin 18 |
| chrM_+_7758 | 0.02 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr8_+_117068582 | 0.01 |
ENSRNOT00000073559
|
Amt
|
aminomethyltransferase |
| chr8_-_6034142 | 0.01 |
ENSRNOT00000014560
|
Birc2
|
baculoviral IAP repeat-containing 2 |
| chr2_-_104133985 | 0.01 |
ENSRNOT00000088167
ENSRNOT00000067725 |
Pde7a
|
phosphodiesterase 7A |
| chr2_+_140836006 | 0.01 |
ENSRNOT00000074141
|
Dusp14l1
|
dual specificity phosphatase 14-like 1 |
| chr3_-_43119159 | 0.01 |
ENSRNOT00000041394
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr19_-_37907714 | 0.01 |
ENSRNOT00000026361
|
Ctrl
|
chymotrypsin-like |
| chr6_-_124429701 | 0.01 |
ENSRNOT00000037902
ENSRNOT00000080950 |
Ttc7b
|
tetratricopeptide repeat domain 7B |
| chr15_-_33250546 | 0.01 |
ENSRNOT00000017857
|
RGD1565222
|
similar to RIKEN cDNA 4931414P19 |
| chr3_+_5624506 | 0.01 |
ENSRNOT00000036995
|
Adamtsl2
|
ADAMTS-like 2 |
| chr18_+_72571327 | 0.01 |
ENSRNOT00000089468
|
Smad2
|
SMAD family member 2 |
| chr20_+_2058197 | 0.01 |
ENSRNOT00000077774
ENSRNOT00000040634 |
RT1-M6-1
|
RT1 class I, locus M6, gene 1 |
| chr13_-_25262469 | 0.01 |
ENSRNOT00000019921
|
Rnf152
|
ring finger protein 152 |
| chr4_+_58053041 | 0.01 |
ENSRNOT00000072698
|
Mest
|
mesoderm specific transcript |
| chr1_+_224824799 | 0.01 |
ENSRNOT00000024757
|
Slc22a6
|
solute carrier family 22 member 6 |
| chr1_-_226353611 | 0.01 |
ENSRNOT00000037624
|
Dagla
|
diacylglycerol lipase, alpha |
| chr11_+_43143882 | 0.01 |
ENSRNOT00000041445
|
Olr1528
|
olfactory receptor 1528 |
| chr10_+_85608544 | 0.01 |
ENSRNOT00000092122
|
Mllt6
|
MLLT6, PHD finger domain containing |
| chr14_+_42714315 | 0.01 |
ENSRNOT00000084095
ENSRNOT00000091449 |
Phox2b
|
paired-like homeobox 2b |
| chr13_-_90467235 | 0.01 |
ENSRNOT00000081534
ENSRNOT00000008150 |
Ncstn
|
nicastrin |
| chr5_+_156587093 | 0.01 |
ENSRNOT00000019696
|
Hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr8_+_36410683 | 0.00 |
ENSRNOT00000015177
|
Srpra
|
SRP receptor alpha subunit |
| chr4_+_71675383 | 0.00 |
ENSRNOT00000051265
|
Clcn1
|
chloride voltage-gated channel 1 |
| chr16_+_70068378 | 0.00 |
ENSRNOT00000073254
|
RGD1308750
|
similar to DNA segment, Chr 19, Brigham & Womens Genetics 1357 expressed |
| chr19_+_39063998 | 0.00 |
ENSRNOT00000081116
|
Has3
|
hyaluronan synthase 3 |
| chr13_-_79801112 | 0.00 |
ENSRNOT00000087323
ENSRNOT00000036483 |
Suco
|
SUN domain containing ossification factor |
| chr18_-_40134504 | 0.00 |
ENSRNOT00000022294
|
Trim36
|
tripartite motif-containing 36 |
| chr10_-_83104173 | 0.00 |
ENSRNOT00000031252
|
Kat7
|
lysine acetyltransferase 7 |
| chr1_-_214423881 | 0.00 |
ENSRNOT00000025290
|
Pidd1
|
p53-induced death domain protein 1 |
| chr8_+_28352772 | 0.00 |
ENSRNOT00000012391
|
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chrX_+_16946207 | 0.00 |
ENSRNOT00000003981
|
RGD1563606
|
similar to cysteine-rich protein 2 |
| chr4_-_82133605 | 0.00 |
ENSRNOT00000008023
|
NEWGENE_2813
|
homeobox A2 |
| chr8_-_36410612 | 0.00 |
ENSRNOT00000091308
|
Foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr18_-_29611212 | 0.00 |
ENSRNOT00000022685
|
Dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr10_-_56511583 | 0.00 |
ENSRNOT00000021402
|
LOC497940
|
similar to RIKEN cDNA 2810408A11 |
| chr2_+_198755262 | 0.00 |
ENSRNOT00000028807
|
Rbm8a
|
RNA binding motif protein 8A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx1_Eomes
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.0 | 0.1 | GO:0090116 | hypermethylation of CpG island(GO:0044027) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.0 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.1 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.0 | GO:0004969 | histamine receptor activity(GO:0004969) |