Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
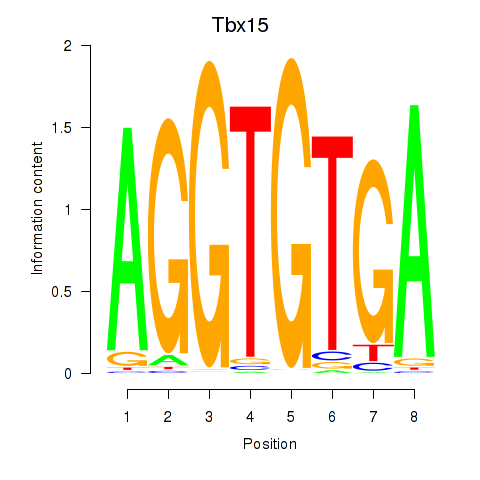
Results for Tbx15
Z-value: 0.54
Transcription factors associated with Tbx15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx15
|
ENSRNOG00000019565 | T-box 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx15 | rn6_v1_chr2_+_201289357_201289357 | 0.49 | 4.0e-01 | Click! |
Activity profile of Tbx15 motif
Sorted Z-values of Tbx15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_45215974 | 0.37 |
ENSRNOT00000010792
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr15_-_30147793 | 0.33 |
ENSRNOT00000060399
|
AABR07017693.1
|
|
| chr5_-_12563429 | 0.29 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr18_+_53609214 | 0.25 |
ENSRNOT00000077443
|
Slc27a6
|
solute carrier family 27 member 6 |
| chr3_+_161433410 | 0.24 |
ENSRNOT00000024657
|
Slc12a5
|
solute carrier family 12 member 5 |
| chr13_+_71107465 | 0.24 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr1_-_264975132 | 0.22 |
ENSRNOT00000021748
|
Lbx1
|
ladybird homeobox 1 |
| chr15_+_8739589 | 0.21 |
ENSRNOT00000071751
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr4_+_140247313 | 0.21 |
ENSRNOT00000040255
ENSRNOT00000064025 ENSRNOT00000041130 ENSRNOT00000043646 |
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr3_+_131351587 | 0.21 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr9_+_9970209 | 0.19 |
ENSRNOT00000075215
|
Dennd1c
|
DENN/MADD domain containing 1C |
| chr10_-_88754829 | 0.18 |
ENSRNOT00000026354
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr1_+_248402980 | 0.18 |
ENSRNOT00000043517
|
Chchd4
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr5_+_143500441 | 0.17 |
ENSRNOT00000045513
|
Grik3
|
glutamate ionotropic receptor kainate type subunit 3 |
| chr10_+_27973681 | 0.16 |
ENSRNOT00000004970
|
Gabrb2
|
gamma-aminobutyric acid type A receptor beta 2 subunit |
| chr10_+_11912543 | 0.16 |
ENSRNOT00000045192
|
Zfp597
|
zinc finger protein 597 |
| chr13_-_25262469 | 0.16 |
ENSRNOT00000019921
|
Rnf152
|
ring finger protein 152 |
| chr1_-_31881531 | 0.15 |
ENSRNOT00000081225
|
Tppp
|
tubulin polymerization promoting protein |
| chr18_-_3662654 | 0.15 |
ENSRNOT00000092854
ENSRNOT00000016167 |
Npc1
|
NPC intracellular cholesterol transporter 1 |
| chr8_+_36625733 | 0.15 |
ENSRNOT00000016248
|
Cdon
|
cell adhesion associated, oncogene regulated |
| chr1_-_219422268 | 0.14 |
ENSRNOT00000025092
|
Carns1
|
carnosine synthase 1 |
| chr1_+_187149453 | 0.13 |
ENSRNOT00000082738
|
Xylt1
|
xylosyltransferase 1 |
| chr10_+_58860940 | 0.13 |
ENSRNOT00000056551
ENSRNOT00000074523 |
XAF1
|
XIAP associated factor-1 |
| chr7_-_107774397 | 0.13 |
ENSRNOT00000093322
|
Ndrg1
|
N-myc downstream regulated 1 |
| chr17_-_10640548 | 0.13 |
ENSRNOT00000064274
|
Simc1
|
SUMO-interacting motifs containing 1 |
| chr12_+_8032809 | 0.12 |
ENSRNOT00000082850
|
Slc7a1
|
solute carrier family 7 member 1 |
| chr5_+_117052260 | 0.12 |
ENSRNOT00000010211
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr1_+_88581206 | 0.12 |
ENSRNOT00000087931
|
Zfp382
|
zinc finger protein 382 |
| chr15_+_34552410 | 0.12 |
ENSRNOT00000027802
|
Khnyn
|
KH and NYN domain containing |
| chr2_-_62634785 | 0.12 |
ENSRNOT00000017937
|
Pdzd2
|
PDZ domain containing 2 |
| chr10_+_76280933 | 0.11 |
ENSRNOT00000051823
|
RGD1304870
|
similar to Tceb2 protein |
| chr10_+_32824 | 0.11 |
ENSRNOT00000077828
|
LOC103689943
|
meiosis arrest female protein 1-like |
| chr12_+_52359310 | 0.11 |
ENSRNOT00000071857
ENSRNOT00000065893 |
Fbrsl1
|
fibrosin-like 1 |
| chr5_-_138697641 | 0.11 |
ENSRNOT00000012062
|
Guca2b
|
guanylate cyclase activator 2B |
| chr19_-_37907714 | 0.11 |
ENSRNOT00000026361
|
Ctrl
|
chymotrypsin-like |
| chr6_-_4520604 | 0.11 |
ENSRNOT00000042230
ENSRNOT00000043870 ENSRNOT00000070918 ENSRNOT00000046246 ENSRNOT00000052367 ENSRNOT00000042251 |
Slc8a1
|
solute carrier family 8 member A1 |
| chr4_-_82133605 | 0.11 |
ENSRNOT00000008023
|
NEWGENE_2813
|
homeobox A2 |
| chr5_-_160742479 | 0.10 |
ENSRNOT00000019447
|
Kazn
|
kazrin, periplakin interacting protein |
| chr16_-_70705128 | 0.10 |
ENSRNOT00000071258
|
Rnf170
|
ring finger protein 170 |
| chr1_-_214423881 | 0.10 |
ENSRNOT00000025290
|
Pidd1
|
p53-induced death domain protein 1 |
| chr1_+_198960542 | 0.10 |
ENSRNOT00000072389
|
Srcap
|
Snf2-related CREBBP activator protein |
| chr15_+_120372 | 0.10 |
ENSRNOT00000007828
|
Dlg5
|
discs large MAGUK scaffold protein 5 |
| chr9_-_63637677 | 0.10 |
ENSRNOT00000049259
|
Satb2
|
SATB homeobox 2 |
| chr10_-_88712309 | 0.10 |
ENSRNOT00000081247
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr3_+_151508361 | 0.10 |
ENSRNOT00000055251
|
Cep250
|
centrosomal protein 250 |
| chr10_+_75149814 | 0.10 |
ENSRNOT00000011500
|
Mks1
|
Meckel syndrome, type 1 |
| chr5_+_136683592 | 0.09 |
ENSRNOT00000085527
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr9_-_81400987 | 0.09 |
ENSRNOT00000035277
|
Tns1
|
tensin 1 |
| chr4_-_117126822 | 0.09 |
ENSRNOT00000086720
|
Rab11fip5
|
RAB11 family interacting protein 5 |
| chr4_+_9981958 | 0.09 |
ENSRNOT00000015322
|
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr16_+_46731403 | 0.09 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr7_-_62972084 | 0.09 |
ENSRNOT00000064251
|
Msrb3
|
methionine sulfoxide reductase B3 |
| chr18_+_79326738 | 0.09 |
ENSRNOT00000089879
|
Mbp
|
myelin basic protein |
| chr14_+_42714315 | 0.09 |
ENSRNOT00000084095
ENSRNOT00000091449 |
Phox2b
|
paired-like homeobox 2b |
| chr10_-_86098694 | 0.09 |
ENSRNOT00000067317
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr20_+_9791171 | 0.09 |
ENSRNOT00000078031
|
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chr2_-_116413873 | 0.09 |
ENSRNOT00000010344
|
Mynn
|
myoneurin |
| chr20_+_2501252 | 0.09 |
ENSRNOT00000079307
ENSRNOT00000084559 |
Trim39
|
tripartite motif-containing 39 |
| chr1_+_238222521 | 0.09 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr4_-_180234804 | 0.08 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr7_-_13108630 | 0.08 |
ENSRNOT00000087038
ENSRNOT00000080491 |
Giot1
|
gonadotropin inducible ovarian transcription factor 1 |
| chr2_-_231521052 | 0.08 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr1_-_90151405 | 0.08 |
ENSRNOT00000028688
|
RGD1308428
|
similar to RIKEN cDNA 4931406P16 |
| chr9_+_92859988 | 0.08 |
ENSRNOT00000085332
|
Cab39
|
calcium binding protein 39 |
| chr11_-_4397361 | 0.08 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chrX_+_111735820 | 0.08 |
ENSRNOT00000086948
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr9_+_10603813 | 0.08 |
ENSRNOT00000073991
ENSRNOT00000074469 ENSRNOT00000079999 |
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr13_-_83425641 | 0.08 |
ENSRNOT00000063870
|
Tbx19
|
T-box 19 |
| chr4_-_78075398 | 0.08 |
ENSRNOT00000010377
|
Zfp467
|
zinc finger protein 467 |
| chr4_+_78024765 | 0.08 |
ENSRNOT00000043856
|
Krba1
|
KRAB-A domain containing 1 |
| chr2_-_177924970 | 0.07 |
ENSRNOT00000029340
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr15_-_33527031 | 0.07 |
ENSRNOT00000019979
|
Homez
|
homeobox and leucine zipper encoding |
| chr17_-_89881919 | 0.07 |
ENSRNOT00000090982
|
LOC100910957
|
acyl-CoA-binding domain-containing protein 5-like |
| chr8_+_98745310 | 0.07 |
ENSRNOT00000019938
|
Zic4
|
Zic family member 4 |
| chr10_-_64642292 | 0.07 |
ENSRNOT00000084670
|
Abr
|
active BCR-related |
| chr10_+_84182118 | 0.07 |
ENSRNOT00000011027
|
Hoxb3
|
homeo box B3 |
| chr2_+_207262934 | 0.07 |
ENSRNOT00000016833
|
Ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr4_+_71675383 | 0.07 |
ENSRNOT00000051265
|
Clcn1
|
chloride voltage-gated channel 1 |
| chr7_+_125940323 | 0.07 |
ENSRNOT00000083830
|
Fam118a
|
family with sequence similarity 118, member A |
| chr8_+_131845696 | 0.07 |
ENSRNOT00000005440
|
Tcaim
|
T cell activation inhibitor, mitochondrial |
| chr11_+_71151132 | 0.07 |
ENSRNOT00000082594
ENSRNOT00000082435 |
Rubcn
|
RUN and cysteine rich domain containing beclin 1 interacting protein |
| chr3_-_80003032 | 0.07 |
ENSRNOT00000017360
|
Madd
|
MAP-kinase activating death domain |
| chr1_+_224824799 | 0.07 |
ENSRNOT00000024757
|
Slc22a6
|
solute carrier family 22 member 6 |
| chr17_-_8429338 | 0.07 |
ENSRNOT00000016390
|
Tgfbi
|
transforming growth factor, beta induced |
| chr3_-_72113680 | 0.07 |
ENSRNOT00000009708
|
Zdhhc5
|
zinc finger, DHHC-type containing 5 |
| chr5_+_117877594 | 0.07 |
ENSRNOT00000011631
|
Atg4c
|
autophagy related 4C, cysteine peptidase |
| chr7_-_12598370 | 0.07 |
ENSRNOT00000026708
|
Arid3a
|
AT-rich interaction domain 3A |
| chr10_-_4257868 | 0.06 |
ENSRNOT00000035886
|
Tnfrsf17
|
TNF receptor superfamily member 17 |
| chr20_+_7431355 | 0.06 |
ENSRNOT00000049592
|
Uhrf1bp1
|
UHRF1 binding protein 1 |
| chr5_-_171710312 | 0.06 |
ENSRNOT00000076044
ENSRNOT00000071280 ENSRNOT00000072059 ENSRNOT00000077015 |
Prdm16
|
PR/SET domain 16 |
| chr10_-_47725172 | 0.06 |
ENSRNOT00000003228
|
Rnf112
|
ring finger protein 112 |
| chr9_+_112360419 | 0.06 |
ENSRNOT00000086682
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr7_+_11383116 | 0.06 |
ENSRNOT00000066348
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr7_-_12598183 | 0.06 |
ENSRNOT00000089009
|
Arid3a
|
AT-rich interaction domain 3A |
| chr10_+_106869401 | 0.06 |
ENSRNOT00000074369
|
Tmem235
|
transmembrane protein 235 |
| chr1_-_214971929 | 0.06 |
ENSRNOT00000027195
|
Mob2
|
MOB kinase activator 2 |
| chr4_+_184019087 | 0.06 |
ENSRNOT00000055435
|
Bicd1
|
BICD cargo adaptor 1 |
| chr1_-_226353611 | 0.06 |
ENSRNOT00000037624
|
Dagla
|
diacylglycerol lipase, alpha |
| chr10_-_73865364 | 0.06 |
ENSRNOT00000005226
|
Rps6kb1
|
ribosomal protein S6 kinase B1 |
| chr18_-_40134504 | 0.06 |
ENSRNOT00000022294
|
Trim36
|
tripartite motif-containing 36 |
| chr1_+_213451721 | 0.06 |
ENSRNOT00000045652
|
Olr315
|
olfactory receptor 315 |
| chr18_-_58097316 | 0.06 |
ENSRNOT00000088527
|
AABR07032265.1
|
|
| chr3_+_63353178 | 0.05 |
ENSRNOT00000091211
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr15_+_83442144 | 0.05 |
ENSRNOT00000039344
|
Bora
|
bora, aurora kinase A activator |
| chr1_+_167051209 | 0.05 |
ENSRNOT00000055249
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr16_-_22411225 | 0.05 |
ENSRNOT00000016204
|
RGD1559708
|
similar to ribosomal protein S10 |
| chr10_-_47018537 | 0.05 |
ENSRNOT00000068351
ENSRNOT00000080083 |
Top3a
|
topoisomerase (DNA) III alpha |
| chr15_+_30429974 | 0.05 |
ENSRNOT00000071797
|
AABR07017735.1
|
|
| chr18_+_61109069 | 0.05 |
ENSRNOT00000068418
|
Malt1
|
MALT1 paracaspase |
| chr2_+_196013799 | 0.05 |
ENSRNOT00000084023
|
Pogz
|
pogo transposable element with ZNF domain |
| chr14_+_83510278 | 0.05 |
ENSRNOT00000081161
|
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr7_-_124198200 | 0.05 |
ENSRNOT00000078947
|
Arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr1_-_142500367 | 0.05 |
ENSRNOT00000015948
ENSRNOT00000089591 |
Crtc3
|
CREB regulated transcription coactivator 3 |
| chr4_+_147037179 | 0.05 |
ENSRNOT00000011292
|
Syn2
|
synapsin II |
| chr10_+_104320743 | 0.05 |
ENSRNOT00000005684
|
Tmem94
|
transmembrane protein 94 |
| chr17_-_10622063 | 0.05 |
ENSRNOT00000022980
|
Simc1
|
SUMO-interacting motifs containing 1 |
| chr10_+_110445797 | 0.05 |
ENSRNOT00000054920
|
Narf
|
nuclear prelamin A recognition factor |
| chr16_-_70665796 | 0.05 |
ENSRNOT00000080051
ENSRNOT00000084974 |
Thap1
|
THAP domain containing 1 |
| chr4_-_56114254 | 0.05 |
ENSRNOT00000010673
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr2_+_29410579 | 0.05 |
ENSRNOT00000023180
|
Zfp366
|
zinc finger protein 366 |
| chr4_+_156752082 | 0.04 |
ENSRNOT00000084588
ENSRNOT00000068407 |
Cd163
|
CD163 molecule |
| chr10_+_61772449 | 0.04 |
ENSRNOT00000004058
|
Smg6
|
SMG6 nonsense mediated mRNA decay factor |
| chr1_+_276309927 | 0.04 |
ENSRNOT00000067460
ENSRNOT00000066236 |
Vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr2_+_74360622 | 0.04 |
ENSRNOT00000014013
|
Cdh18
|
cadherin 18 |
| chr8_-_82533689 | 0.04 |
ENSRNOT00000014124
|
Tmod2
|
tropomodulin 2 |
| chr5_+_76386836 | 0.04 |
ENSRNOT00000088872
ENSRNOT00000021110 |
Ugcg
|
UDP-glucose ceramide glucosyltransferase |
| chr13_-_47154292 | 0.04 |
ENSRNOT00000005284
|
Cd55
|
CD55 molecule, decay accelerating factor for complement |
| chr16_-_82099991 | 0.04 |
ENSRNOT00000089447
|
Atp11a
|
ATPase phospholipid transporting 11A |
| chr8_-_49158971 | 0.04 |
ENSRNOT00000020573
|
Kmt2a
|
lysine methyltransferase 2A |
| chr14_+_39663421 | 0.04 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr2_+_120266933 | 0.04 |
ENSRNOT00000015192
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr4_-_55527092 | 0.04 |
ENSRNOT00000087407
ENSRNOT00000010223 |
Zfp800
|
zinc finger protein 800 |
| chr9_+_27554751 | 0.04 |
ENSRNOT00000077690
|
AABR07067042.3
|
|
| chr6_+_80108655 | 0.04 |
ENSRNOT00000006137
|
Gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr10_+_63778755 | 0.04 |
ENSRNOT00000090481
|
Inpp5k
|
inositol polyphosphate-5-phosphatase K |
| chr6_+_72092194 | 0.04 |
ENSRNOT00000005670
|
G2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr3_+_16846412 | 0.04 |
ENSRNOT00000074266
|
AABR07051551.1
|
|
| chr6_+_28323647 | 0.04 |
ENSRNOT00000089093
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr2_-_27364906 | 0.04 |
ENSRNOT00000078639
|
Polk
|
DNA polymerase kappa |
| chr10_-_57121584 | 0.04 |
ENSRNOT00000029421
|
Vmo1
|
vitelline membrane outer layer 1 homolog |
| chr9_-_61810417 | 0.04 |
ENSRNOT00000020910
|
Rftn2
|
raftlin family member 2 |
| chr16_+_20039969 | 0.04 |
ENSRNOT00000024866
|
Colgalt1
|
collagen beta(1-O)galactosyltransferase 1 |
| chr8_+_116826680 | 0.04 |
ENSRNOT00000026854
|
Gmppb
|
GDP-mannose pyrophosphorylase B |
| chr15_+_18941431 | 0.04 |
ENSRNOT00000092092
|
AABR07017236.1
|
|
| chr19_-_54722563 | 0.04 |
ENSRNOT00000025784
|
Slc7a5
|
solute carrier family 7 member 5 |
| chr3_-_176744377 | 0.04 |
ENSRNOT00000017787
|
Helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr7_-_139978921 | 0.04 |
ENSRNOT00000048039
|
Olr1106
|
olfactory receptor 1106 |
| chr20_+_34633157 | 0.04 |
ENSRNOT00000000469
|
Pln
|
phospholamban |
| chr3_-_151625644 | 0.03 |
ENSRNOT00000026657
|
Rbm12
|
RNA binding motif protein 12 |
| chr11_+_61662270 | 0.03 |
ENSRNOT00000079521
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr3_+_80556668 | 0.03 |
ENSRNOT00000079118
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr1_-_81152384 | 0.03 |
ENSRNOT00000026263
ENSRNOT00000077305 |
Zfp94
|
zinc finger protein 94 |
| chr10_-_61772250 | 0.03 |
ENSRNOT00000092338
ENSRNOT00000085394 ENSRNOT00000046110 |
Srr
|
serine racemase |
| chr1_-_81163689 | 0.03 |
ENSRNOT00000088746
|
LOC102548695
|
zinc finger protein 45-like |
| chr6_-_44363915 | 0.03 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr4_+_109467272 | 0.03 |
ENSRNOT00000008212
|
Reg3b
|
regenerating family member 3 beta |
| chr2_+_143895982 | 0.03 |
ENSRNOT00000080026
|
Supt20h
|
SPT20 homolog, SAGA complex component |
| chr14_+_1462358 | 0.03 |
ENSRNOT00000077243
|
Csf2ra
|
colony stimulating factor 2 receptor alpha subunit |
| chr12_+_25430464 | 0.03 |
ENSRNOT00000002020
|
Gtf2i
|
general transcription factor II I |
| chr12_+_49419582 | 0.03 |
ENSRNOT00000082998
|
RGD1306556
|
similar to hypothetical protein A530094D01 |
| chrX_-_158925630 | 0.03 |
ENSRNOT00000073396
|
Ct45a9
|
cancer/testis antigen family 45 member A9 |
| chr1_-_260517323 | 0.03 |
ENSRNOT00000018043
|
Tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr15_+_31950986 | 0.03 |
ENSRNOT00000080233
|
AABR07017868.4
|
|
| chr1_+_88542035 | 0.03 |
ENSRNOT00000032973
|
Zfp420
|
zinc finger protein 420 |
| chr13_+_56513286 | 0.03 |
ENSRNOT00000015596
|
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chrX_-_65102344 | 0.03 |
ENSRNOT00000016042
ENSRNOT00000075875 |
Las1l
|
LAS1-like, ribosome biogenesis factor |
| chr2_+_60920257 | 0.03 |
ENSRNOT00000025170
|
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr4_-_78074906 | 0.03 |
ENSRNOT00000080654
|
Zfp467
|
zinc finger protein 467 |
| chr1_+_273854248 | 0.03 |
ENSRNOT00000044827
ENSRNOT00000017600 ENSRNOT00000086496 |
Add3
|
adducin 3 |
| chr11_+_80255790 | 0.03 |
ENSRNOT00000002522
|
Bcl6
|
B-cell CLL/lymphoma 6 |
| chr6_+_101532518 | 0.02 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr19_-_25288335 | 0.02 |
ENSRNOT00000050214
|
Nanos3
|
nanos C2HC-type zinc finger 3 |
| chr16_-_48437223 | 0.02 |
ENSRNOT00000013005
ENSRNOT00000059401 |
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr10_+_85608544 | 0.02 |
ENSRNOT00000092122
|
Mllt6
|
MLLT6, PHD finger domain containing |
| chr17_+_1679627 | 0.02 |
ENSRNOT00000025801
|
Habp4
|
hyaluronan binding protein 4 |
| chr9_-_14668297 | 0.02 |
ENSRNOT00000042404
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr2_+_140541619 | 0.02 |
ENSRNOT00000017396
|
Rab33b
|
RAB33B, member RAS oncogene family |
| chr19_+_37600148 | 0.02 |
ENSRNOT00000023853
|
Ctcf
|
CCCTC-binding factor |
| chr1_+_88324339 | 0.02 |
ENSRNOT00000072762
|
LOC102547059
|
zinc finger protein 420-like |
| chr7_+_143679617 | 0.02 |
ENSRNOT00000014033
|
Eif4b
|
eukaryotic translation initiation factor 4B |
| chr8_-_78233430 | 0.02 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chrX_+_65563122 | 0.02 |
ENSRNOT00000017312
|
Heph
|
hephaestin |
| chr9_+_41045363 | 0.02 |
ENSRNOT00000066284
|
Cfc1
|
cripto, FRL-1, cryptic family 1 |
| chr13_-_79801112 | 0.02 |
ENSRNOT00000087323
ENSRNOT00000036483 |
Suco
|
SUN domain containing ossification factor |
| chr1_+_87903783 | 0.02 |
ENSRNOT00000074299
|
Zfp84
|
zinc finger protein 84 |
| chr6_-_77243677 | 0.02 |
ENSRNOT00000011419
|
Mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr5_-_147412705 | 0.02 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr1_-_72468738 | 0.02 |
ENSRNOT00000029136
|
Zfp628
|
zinc finger protein 628 |
| chr10_-_57309298 | 0.02 |
ENSRNOT00000056622
|
Camta2
|
calmodulin binding transcription activator 2 |
| chr5_-_9432343 | 0.02 |
ENSRNOT00000009482
|
Rrs1
|
ribosome biogenesis regulator homolog |
| chr1_-_265573117 | 0.02 |
ENSRNOT00000044195
ENSRNOT00000055915 |
LOC100911951
|
Kv channel-interacting protein 2-like |
| chr7_-_3074359 | 0.02 |
ENSRNOT00000019738
|
Ikzf4
|
IKAROS family zinc finger 4 |
| chr20_-_45126062 | 0.02 |
ENSRNOT00000000720
|
RGD1310495
|
similar to KIAA1919 protein |
| chr7_-_123767797 | 0.02 |
ENSRNOT00000012699
|
Tcf20
|
transcription factor 20 |
| chr5_+_145145983 | 0.02 |
ENSRNOT00000079732
|
Zmym6
|
zinc finger MYM-type containing 6 |
| chr10_-_47857326 | 0.02 |
ENSRNOT00000085703
ENSRNOT00000091963 |
Epn2
|
epsin 2 |
| chr6_+_36107672 | 0.02 |
ENSRNOT00000093003
|
Rdh14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
| chr17_+_11856525 | 0.02 |
ENSRNOT00000014546
|
Sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr4_+_106323089 | 0.02 |
ENSRNOT00000091402
|
AABR07061134.1
|
|
| chr10_-_72196437 | 0.02 |
ENSRNOT00000029769
|
Pigw
|
phosphatidylinositol glycan anchor biosynthesis, class W |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx15
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.1 | 0.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.4 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.0 | 0.1 | GO:0021550 | medulla oblongata development(GO:0021550) sensory system development(GO:0048880) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.1 | GO:0036395 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) retrograde trans-synaptic signaling(GO:0098917) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:1990822 | arginine transmembrane transport(GO:1903826) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.0 | GO:2001038 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) regulation of cellular response to drug(GO:2001038) |
| 0.0 | 0.1 | GO:0031427 | response to methotrexate(GO:0031427) |
| 0.0 | 0.0 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.0 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.0 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.1 | GO:0042905 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.0 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.0 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.0 | 0.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.0 | GO:0090534 | calcium ion-transporting ATPase complex(GO:0090534) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.0 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |