Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Tbr1
Z-value: 0.84
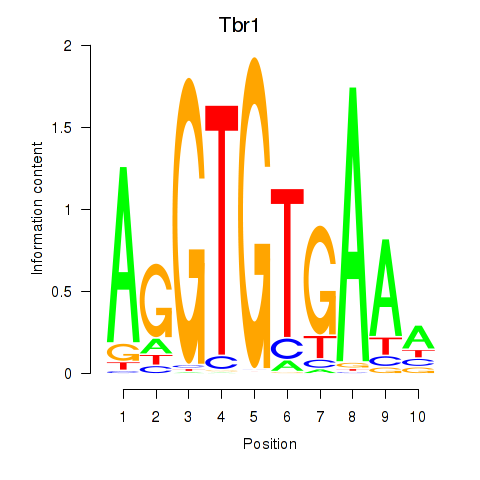
Transcription factors associated with Tbr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbr1
|
ENSRNOG00000005049 | T-box, brain, 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbr1 | rn6_v1_chr3_+_47677720_47677720 | 0.13 | 8.3e-01 | Click! |
Activity profile of Tbr1 motif
Sorted Z-values of Tbr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_109300433 | 0.71 |
ENSRNOT00000010712
|
Fos
|
FBJ osteosarcoma oncogene |
| chr7_-_82059247 | 0.36 |
ENSRNOT00000087177
|
Rspo2
|
R-spondin 2 |
| chr1_+_219250265 | 0.34 |
ENSRNOT00000024353
|
Doc2g
|
double C2-like domains, gamma |
| chr14_+_63405408 | 0.30 |
ENSRNOT00000086658
|
AABR07015559.1
|
|
| chr1_-_169513537 | 0.30 |
ENSRNOT00000078058
|
Trim30c
|
tripartite motif-containing 30C |
| chr10_+_14136883 | 0.27 |
ENSRNOT00000082295
|
AC115181.1
|
|
| chr6_-_115513354 | 0.27 |
ENSRNOT00000005881
|
Ston2
|
stonin 2 |
| chr1_-_82004538 | 0.27 |
ENSRNOT00000087572
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr5_-_74029238 | 0.26 |
ENSRNOT00000031432
|
Frrs1l
|
ferric-chelate reductase 1-like |
| chr6_+_106039991 | 0.26 |
ENSRNOT00000088917
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr1_-_25839198 | 0.26 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr1_-_199655147 | 0.25 |
ENSRNOT00000026979
|
LOC103691238
|
zinc finger protein 239-like |
| chr3_-_43119159 | 0.25 |
ENSRNOT00000041394
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr2_+_251200686 | 0.25 |
ENSRNOT00000019210
|
Col24a1
|
collagen type XXIV alpha 1 chain |
| chr9_-_20528879 | 0.24 |
ENSRNOT00000085293
|
AABR07066871.3
|
|
| chr20_-_22459025 | 0.24 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chr8_+_58431407 | 0.24 |
ENSRNOT00000011974
|
Sln
|
sarcolipin |
| chr7_-_144936803 | 0.23 |
ENSRNOT00000055279
|
Gpr84
|
G protein-coupled receptor 84 |
| chr9_-_81566642 | 0.23 |
ENSRNOT00000080345
|
Aamp
|
angio-associated, migratory cell protein |
| chr13_-_61591139 | 0.22 |
ENSRNOT00000005324
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr18_+_25613831 | 0.22 |
ENSRNOT00000091040
|
Tslp
|
thymic stromal lymphopoietin |
| chr10_+_64737022 | 0.21 |
ENSRNOT00000017071
ENSRNOT00000093232 ENSRNOT00000017042 ENSRNOT00000093244 |
Lgals9
|
galectin 9 |
| chr7_-_123892429 | 0.21 |
ENSRNOT00000037681
|
Nfam1
|
NFAT activating protein with ITAM motif 1 |
| chr8_-_64305330 | 0.20 |
ENSRNOT00000030199
|
Tmem202
|
transmembrane protein 202 |
| chr1_+_142679345 | 0.20 |
ENSRNOT00000034267
|
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr5_+_74727494 | 0.20 |
ENSRNOT00000076683
|
Rn50_5_0791.1
|
|
| chr15_+_62406873 | 0.20 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr1_-_219168177 | 0.20 |
ENSRNOT00000023789
|
Aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr5_-_136098013 | 0.20 |
ENSRNOT00000089166
|
RGD1563714
|
RGD1563714 |
| chr13_+_101790865 | 0.20 |
ENSRNOT00000087784
|
Taf1a
|
TATA-box binding protein associated factor, RNA polymerase I subunit A |
| chr2_+_140471690 | 0.20 |
ENSRNOT00000017379
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr8_-_45137893 | 0.19 |
ENSRNOT00000010743
|
RGD1309108
|
similar to hypothetical protein FLJ23554 |
| chr1_-_227047290 | 0.19 |
ENSRNOT00000037226
|
Ms4a15
|
membrane spanning 4-domains A15 |
| chrX_+_13117239 | 0.19 |
ENSRNOT00000088998
|
AABR07037112.1
|
|
| chr1_+_170242846 | 0.19 |
ENSRNOT00000023751
|
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr1_+_80777014 | 0.19 |
ENSRNOT00000079758
|
Gm19345
|
predicted gene, 19345 |
| chr10_+_11912543 | 0.19 |
ENSRNOT00000045192
|
Zfp597
|
zinc finger protein 597 |
| chr16_-_32868680 | 0.19 |
ENSRNOT00000015974
ENSRNOT00000082392 |
Aadat
|
aminoadipate aminotransferase |
| chr20_+_4043741 | 0.18 |
ENSRNOT00000000525
|
RT1-Bb
|
RT1 class II, locus Bb |
| chr11_-_81444375 | 0.18 |
ENSRNOT00000058479
ENSRNOT00000078131 ENSRNOT00000080949 ENSRNOT00000080562 ENSRNOT00000084867 |
Kng1
|
kininogen 1 |
| chr1_+_161401527 | 0.18 |
ENSRNOT00000015181
|
Tenm4
|
teneurin transmembrane protein 4 |
| chr10_-_87541851 | 0.18 |
ENSRNOT00000089610
|
RGD1561684
|
similar to keratin associated protein 2-4 |
| chr3_+_15560712 | 0.18 |
ENSRNOT00000010218
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr9_-_15700235 | 0.18 |
ENSRNOT00000088713
ENSRNOT00000035907 |
Trerf1
|
transcriptional regulating factor 1 |
| chr8_+_33239139 | 0.17 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr5_+_164808323 | 0.17 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr12_-_41448668 | 0.17 |
ENSRNOT00000001856
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr14_-_92495894 | 0.17 |
ENSRNOT00000064483
|
Cobl
|
cordon-bleu WH2 repeat protein |
| chr5_-_106207126 | 0.17 |
ENSRNOT00000015094
|
Mllt3
|
MLLT3, super elongation complex subunit |
| chr15_-_42898150 | 0.16 |
ENSRNOT00000030036
|
Ptk2b
|
protein tyrosine kinase 2 beta |
| chr7_+_64769089 | 0.16 |
ENSRNOT00000088861
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr4_+_171250818 | 0.16 |
ENSRNOT00000040576
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr12_-_9990284 | 0.16 |
ENSRNOT00000001264
|
Rasl11a
|
RAS-like family 11 member A |
| chr17_-_10622063 | 0.16 |
ENSRNOT00000022980
|
Simc1
|
SUMO-interacting motifs containing 1 |
| chr2_-_25153334 | 0.16 |
ENSRNOT00000024191
|
Crhbp
|
corticotropin releasing hormone binding protein |
| chr7_+_121841855 | 0.15 |
ENSRNOT00000024673
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr7_-_80457816 | 0.15 |
ENSRNOT00000039430
|
AABR07057617.1
|
|
| chr15_+_1054937 | 0.15 |
ENSRNOT00000008154
|
AABR07016841.1
|
|
| chr19_-_56037077 | 0.15 |
ENSRNOT00000021968
|
Spata2L
|
spermatogenesis associated 2-like |
| chr13_+_50103189 | 0.15 |
ENSRNOT00000004078
|
Atp2b4
|
ATPase plasma membrane Ca2+ transporting 4 |
| chr14_+_12218553 | 0.15 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr9_-_104350308 | 0.15 |
ENSRNOT00000033958
|
Slco4c1
|
solute carrier organic anion transporter family, member 4C1 |
| chr4_-_51586881 | 0.15 |
ENSRNOT00000064983
|
Iqub
|
IQ motif and ubiquitin domain containing |
| chr1_+_42169501 | 0.15 |
ENSRNOT00000025477
ENSRNOT00000092791 |
Vip
|
vasoactive intestinal peptide |
| chr10_+_1622573 | 0.15 |
ENSRNOT00000040132
ENSRNOT00000090207 ENSRNOT00000089118 |
AABR07028989.1
|
|
| chr19_+_39063998 | 0.14 |
ENSRNOT00000081116
|
Has3
|
hyaluronan synthase 3 |
| chr13_+_50974872 | 0.14 |
ENSRNOT00000080768
|
Chit1
|
chitinase 1 |
| chr10_-_57121584 | 0.14 |
ENSRNOT00000029421
|
Vmo1
|
vitelline membrane outer layer 1 homolog |
| chr10_-_74119009 | 0.14 |
ENSRNOT00000085712
ENSRNOT00000006926 |
Dhx40
|
DEAH-box helicase 40 |
| chr15_+_83442144 | 0.14 |
ENSRNOT00000039344
|
Bora
|
bora, aurora kinase A activator |
| chr1_+_196996581 | 0.14 |
ENSRNOT00000021690
|
Il21r
|
interleukin 21 receptor |
| chr8_+_29453643 | 0.14 |
ENSRNOT00000090643
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr6_-_25616995 | 0.14 |
ENSRNOT00000077894
|
Fosl2
|
FOS like 2, AP-1 transcription factor subunit |
| chr13_-_50509916 | 0.14 |
ENSRNOT00000076747
|
Ren
|
renin |
| chr1_-_155955173 | 0.14 |
ENSRNOT00000079345
|
AABR07004776.1
|
|
| chr17_-_1610745 | 0.14 |
ENSRNOT00000025850
|
Hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr1_+_84584596 | 0.14 |
ENSRNOT00000032933
|
LOC100910046
|
zinc finger protein 60-like |
| chr4_-_124338176 | 0.14 |
ENSRNOT00000016628
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr4_+_2053712 | 0.14 |
ENSRNOT00000045086
|
Rnf32
|
ring finger protein 32 |
| chr7_+_48867664 | 0.14 |
ENSRNOT00000005862
|
Ppfia2
|
PTPRF interacting protein alpha 2 |
| chr18_-_37096132 | 0.13 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr12_+_2534212 | 0.13 |
ENSRNOT00000001399
|
Ctxn1
|
cortexin 1 |
| chr4_+_158224000 | 0.13 |
ENSRNOT00000084240
ENSRNOT00000078495 |
Ano2
|
anoctamin 2 |
| chr7_+_15422479 | 0.13 |
ENSRNOT00000066520
|
Zfp563
|
zinc finger protein 563 |
| chr3_+_71437241 | 0.13 |
ENSRNOT00000046112
|
LOC499823
|
LRRG00114 |
| chr10_-_91661558 | 0.13 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
| chr7_-_104801045 | 0.13 |
ENSRNOT00000079524
|
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr1_-_261446570 | 0.13 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chrX_-_142248369 | 0.13 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr18_+_68984051 | 0.13 |
ENSRNOT00000002969
|
Stard6
|
StAR-related lipid transfer domain containing 6 |
| chrX_-_70428364 | 0.13 |
ENSRNOT00000045907
|
P2ry4
|
pyrimidinergic receptor P2Y4 |
| chr13_+_21678512 | 0.13 |
ENSRNOT00000047108
|
Cntnap5b
|
contactin associated protein-like 5B |
| chr1_-_82003691 | 0.13 |
ENSRNOT00000084569
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr13_-_89661150 | 0.13 |
ENSRNOT00000058390
|
Usp21
|
ubiquitin specific peptidase 21 |
| chr4_+_62299044 | 0.13 |
ENSRNOT00000032077
|
Agbl3
|
ATP/GTP binding protein-like 3 |
| chr13_+_80464348 | 0.13 |
ENSRNOT00000076324
|
Vamp4
|
vesicle-associated membrane protein 4 |
| chr20_+_14578605 | 0.13 |
ENSRNOT00000041165
|
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr17_+_10066187 | 0.12 |
ENSRNOT00000091200
|
Uimc1
|
ubiquitin interaction motif containing 1 |
| chr1_+_21613148 | 0.12 |
ENSRNOT00000018695
|
Enpp3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr16_+_39909270 | 0.12 |
ENSRNOT00000081994
|
Wdr17
|
WD repeat domain 17 |
| chr20_-_45126062 | 0.12 |
ENSRNOT00000000720
|
RGD1310495
|
similar to KIAA1919 protein |
| chr5_+_2654065 | 0.12 |
ENSRNOT00000074957
|
LOC100912291
|
uncharacterized LOC100912291 |
| chr4_+_78371121 | 0.12 |
ENSRNOT00000059157
|
Gimap1
|
GTPase, IMAP family member 1 |
| chr1_-_42486035 | 0.12 |
ENSRNOT00000025398
|
Mtrf1l
|
mitochondrial translational release factor 1-like |
| chr5_+_58661049 | 0.12 |
ENSRNOT00000078274
|
Unc13b
|
unc-13 homolog B |
| chr2_-_19808937 | 0.12 |
ENSRNOT00000044237
|
Atp6ap1l
|
ATPase H+ transporting accessory protein 1 like |
| chr14_-_108658371 | 0.12 |
ENSRNOT00000008919
|
Papolg
|
poly(A) polymerase gamma |
| chr13_-_103080920 | 0.12 |
ENSRNOT00000034990
|
AABR07022022.1
|
|
| chr1_+_248647170 | 0.12 |
ENSRNOT00000015016
|
Tpd52l3
|
tumor protein D52-like 3 |
| chr3_-_6626284 | 0.12 |
ENSRNOT00000012494
|
Fcnb
|
ficolin B |
| chr17_-_21739408 | 0.12 |
ENSRNOT00000060335
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr1_+_88875375 | 0.12 |
ENSRNOT00000028284
|
Tyrobp
|
Tyro protein tyrosine kinase binding protein |
| chr20_-_6500523 | 0.12 |
ENSRNOT00000000629
|
Cpne5
|
copine 5 |
| chr1_-_97785490 | 0.12 |
ENSRNOT00000019364
|
LOC499136
|
LRRGT00021 |
| chr1_+_87938042 | 0.12 |
ENSRNOT00000027837
|
Map4k1
|
mitogen activated protein kinase kinase kinase kinase 1 |
| chr13_-_74331214 | 0.12 |
ENSRNOT00000006018
|
Fam20b
|
FAM20B, glycosaminoglycan xylosylkinase |
| chr3_+_72080630 | 0.12 |
ENSRNOT00000008911
|
Med19
|
mediator complex subunit 19 |
| chr9_+_61738471 | 0.12 |
ENSRNOT00000090305
|
AABR07067762.1
|
|
| chr4_+_181103774 | 0.12 |
ENSRNOT00000084207
ENSRNOT00000055473 ENSRNOT00000077619 |
Arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr1_+_47942800 | 0.12 |
ENSRNOT00000079312
|
Wtap
|
Wilms tumor 1 associated protein |
| chr7_-_70498992 | 0.12 |
ENSRNOT00000067774
ENSRNOT00000079327 |
Pip4k2c
|
phosphatidylinositol-5-phosphate 4-kinase type 2 gamma |
| chr4_-_118342176 | 0.11 |
ENSRNOT00000032477
|
AC139642.2
|
|
| chr2_-_23289266 | 0.11 |
ENSRNOT00000061708
|
Bhmt2
|
betaine-homocysteine S-methyltransferase 2 |
| chr1_+_73837944 | 0.11 |
ENSRNOT00000036413
|
Lair1
|
leukocyte-associated immunoglobulin-like receptor 1 |
| chr7_-_104800536 | 0.11 |
ENSRNOT00000085650
|
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr13_+_48644604 | 0.11 |
ENSRNOT00000084259
ENSRNOT00000073669 |
Rab29
|
RAB29, member RAS oncogene family |
| chr11_-_32508420 | 0.11 |
ENSRNOT00000002717
|
Kcne1
|
potassium voltage-gated channel subfamily E regulatory subunit 1 |
| chr8_+_113105814 | 0.11 |
ENSRNOT00000016749
|
Cpne4
|
copine 4 |
| chr8_-_83280888 | 0.11 |
ENSRNOT00000052341
|
Gfral
|
GDNF family receptor alpha like |
| chr8_-_22336794 | 0.11 |
ENSRNOT00000066340
|
Ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr6_+_107517668 | 0.11 |
ENSRNOT00000013753
|
Acot4
|
acyl-CoA thioesterase 4 |
| chr1_+_48077033 | 0.11 |
ENSRNOT00000020100
|
Mas1
|
MAS1 proto-oncogene, G protein-coupled receptor |
| chr2_+_55835151 | 0.11 |
ENSRNOT00000018634
|
Fyb
|
FYN binding protein |
| chr4_+_114854458 | 0.11 |
ENSRNOT00000013312
|
LOC500227
|
hypothetical gene supported by BC079424 |
| chr10_+_1920529 | 0.11 |
ENSRNOT00000072296
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chr3_+_100768637 | 0.11 |
ENSRNOT00000083542
|
Bdnf
|
brain-derived neurotrophic factor |
| chr3_-_153321352 | 0.11 |
ENSRNOT00000064824
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chr9_+_12114977 | 0.11 |
ENSRNOT00000073673
|
AABR07066648.1
|
|
| chr2_-_52492509 | 0.11 |
ENSRNOT00000021964
|
Nim1k
|
NIM1 serine/threonine protein kinase |
| chr18_-_44141865 | 0.11 |
ENSRNOT00000083407
|
LOC103694210
|
NACHT and WD repeat domain-containing protein 2-like |
| chr2_-_106703401 | 0.11 |
ENSRNOT00000043933
|
AABR07009638.1
|
|
| chr2_-_211322719 | 0.11 |
ENSRNOT00000027493
|
RGD1310209
|
similar to KIAA1324 protein |
| chr5_+_104698040 | 0.11 |
ENSRNOT00000009127
|
Adamtsl1
|
ADAMTS-like 1 |
| chr7_+_143096874 | 0.11 |
ENSRNOT00000010881
|
RGD1305207
|
similar to RIKEN cDNA 1700011A15 |
| chr7_-_97994586 | 0.11 |
ENSRNOT00000077616
|
Wdyhv1
|
WDYHV motif containing 1 |
| chr13_+_113691932 | 0.11 |
ENSRNOT00000084709
|
Cd34
|
CD34 molecule |
| chr3_+_48626038 | 0.10 |
ENSRNOT00000009697
|
Gca
|
grancalcin |
| chr15_-_33629699 | 0.10 |
ENSRNOT00000023302
|
Myh6
|
myosin heavy chain 6 |
| chr10_+_69423086 | 0.10 |
ENSRNOT00000000256
|
Ccl7
|
C-C motif chemokine ligand 7 |
| chr3_-_123849788 | 0.10 |
ENSRNOT00000028869
|
Rnf24
|
ring finger protein 24 |
| chr1_+_105094411 | 0.10 |
ENSRNOT00000036258
|
Htatip2
|
HIV-1 Tat interactive protein 2 |
| chr13_-_70626252 | 0.10 |
ENSRNOT00000036947
|
Lamc2
|
laminin subunit gamma 2 |
| chr20_+_28572242 | 0.10 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr2_-_21698937 | 0.10 |
ENSRNOT00000080165
|
AABR07007642.1
|
|
| chr10_+_53570989 | 0.10 |
ENSRNOT00000064764
ENSRNOT00000004516 |
Tmem220
|
transmembrane protein 220 |
| chr2_-_198834038 | 0.10 |
ENSRNOT00000031484
|
Nudt17
|
nudix hydrolase 17 |
| chr9_-_30251388 | 0.10 |
ENSRNOT00000035033
|
Sdhaf4
|
succinate dehydrogenase complex assembly factor 4 |
| chr1_-_151106802 | 0.10 |
ENSRNOT00000021971
|
Tyr
|
tyrosinase |
| chr1_-_72377434 | 0.10 |
ENSRNOT00000022193
|
Zfp524
|
zinc finger protein 524 |
| chr20_-_3401273 | 0.10 |
ENSRNOT00000089257
ENSRNOT00000078451 ENSRNOT00000001085 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr6_+_78567970 | 0.10 |
ENSRNOT00000032743
|
Ttc6
|
tetratricopeptide repeat domain 6 |
| chr7_+_41475163 | 0.10 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr3_+_159321310 | 0.10 |
ENSRNOT00000045677
ENSRNOT00000063906 |
L3mbtl1
|
l(3)mbt-like 1 (Drosophila) |
| chr5_+_59228199 | 0.10 |
ENSRNOT00000060282
|
Hrct1
|
histidine rich carboxyl terminus 1 |
| chr1_-_71290337 | 0.10 |
ENSRNOT00000063956
|
Zfp667
|
zinc finger protein 667 |
| chr14_-_3462629 | 0.10 |
ENSRNOT00000061538
|
Brdt
|
bromodomain testis associated |
| chr1_+_205706468 | 0.10 |
ENSRNOT00000089957
ENSRNOT00000023877 |
Edrf1
|
erythroid differentiation regulatory factor 1 |
| chr3_-_67668772 | 0.10 |
ENSRNOT00000010247
|
Frzb
|
frizzled-related protein |
| chr2_-_210766501 | 0.10 |
ENSRNOT00000025800
|
Gstm3
|
glutathione S-transferase mu 3 |
| chr11_+_27208564 | 0.10 |
ENSRNOT00000002158
|
Map3k7cl
|
MAP3K7 C-terminal like |
| chr7_+_23960378 | 0.10 |
ENSRNOT00000006886
|
Prdm4
|
PR/SET domain 4 |
| chr10_-_68926264 | 0.10 |
ENSRNOT00000008661
|
Phb-ps1
|
prohibitin, pseudogene 1 |
| chr12_-_31629881 | 0.09 |
ENSRNOT00000001238
|
Piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr15_+_27739251 | 0.09 |
ENSRNOT00000011840
|
Parp2
|
poly (ADP-ribose) polymerase 2 |
| chr6_-_50923510 | 0.09 |
ENSRNOT00000010631
|
Bcap29
|
B-cell receptor-associated protein 29 |
| chr10_-_73865364 | 0.09 |
ENSRNOT00000005226
|
Rps6kb1
|
ribosomal protein S6 kinase B1 |
| chr10_-_103685844 | 0.09 |
ENSRNOT00000064284
|
Cd300lf
|
Cd300 molecule-like family member F |
| chrX_+_120859968 | 0.09 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr7_-_12899004 | 0.09 |
ENSRNOT00000011086
|
Gzmm
|
granzyme M |
| chr16_-_84939323 | 0.09 |
ENSRNOT00000022742
|
Myo16
|
myosin XVI |
| chrX_+_16946207 | 0.09 |
ENSRNOT00000003981
|
RGD1563606
|
similar to cysteine-rich protein 2 |
| chrX_+_104734082 | 0.09 |
ENSRNOT00000005020
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr7_+_14891001 | 0.09 |
ENSRNOT00000041034
|
Cyp4f4
|
cytochrome P450, family 4, subfamily f, polypeptide 4 |
| chr3_-_36660758 | 0.09 |
ENSRNOT00000006111
|
Rnd3
|
Rho family GTPase 3 |
| chr18_+_69549937 | 0.09 |
ENSRNOT00000041227
|
Mex3c
|
mex-3 RNA binding family member C |
| chr2_+_58448917 | 0.09 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chr15_+_48674380 | 0.09 |
ENSRNOT00000018762
|
Fbxo16
|
F-box protein 16 |
| chr1_+_266333440 | 0.09 |
ENSRNOT00000071243
|
Sfxn2
|
sideroflexin 2 |
| chr7_+_116355698 | 0.09 |
ENSRNOT00000076693
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr1_+_89220083 | 0.09 |
ENSRNOT00000093144
|
Dmkn
|
dermokine |
| chr19_-_11341863 | 0.09 |
ENSRNOT00000025694
|
Mt4
|
metallothionein 4 |
| chr7_+_34326087 | 0.09 |
ENSRNOT00000006971
|
Hal
|
histidine ammonia lyase |
| chrX_-_68562873 | 0.09 |
ENSRNOT00000076193
|
Ophn1
|
oligophrenin 1 |
| chr7_-_120744602 | 0.09 |
ENSRNOT00000018564
|
Kcnj4
|
potassium voltage-gated channel subfamily J member 4 |
| chr1_-_128695995 | 0.09 |
ENSRNOT00000077020
|
Synm
|
synemin |
| chr1_+_88581206 | 0.09 |
ENSRNOT00000087931
|
Zfp382
|
zinc finger protein 382 |
| chr17_-_21705773 | 0.09 |
ENSRNOT00000078010
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr17_+_9595761 | 0.09 |
ENSRNOT00000089990
|
Fam193b
|
family with sequence similarity 193, member B |
| chr19_+_9622611 | 0.09 |
ENSRNOT00000061498
|
Slc38a7
|
solute carrier family 38, member 7 |
| chr1_+_248195797 | 0.09 |
ENSRNOT00000066891
|
LOC103690131
|
tumor protein D55-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.1 | 0.4 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.1 | 0.9 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.3 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.3 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.1 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 0.3 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.2 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.2 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.1 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.2 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.1 | 0.2 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.3 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0017143 | insecticide metabolic process(GO:0017143) |
| 0.0 | 0.1 | GO:2000041 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:2001038 | regulation of cellular response to drug(GO:2001038) |
| 0.0 | 0.1 | GO:1905243 | response to 3,3',5-triiodo-L-thyronine(GO:1905242) cellular response to 3,3',5-triiodo-L-thyronine(GO:1905243) |
| 0.0 | 0.1 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) |
| 0.0 | 0.2 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:1900106 | hyaluranon cable assembly(GO:0036118) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.2 | GO:0048669 | somite specification(GO:0001757) collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:2000426 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.1 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.1 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:1902024 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.1 | GO:0002018 | angiotensin maturation(GO:0002003) renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.2 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) renal artery morphogenesis(GO:0061441) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.0 | 0.2 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.1 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.1 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.1 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0032639 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:2000541 | positive regulation of synaptic growth at neuromuscular junction(GO:0045887) regulation of protein geranylgeranylation(GO:2000539) positive regulation of protein geranylgeranylation(GO:2000541) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:2000118 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.1 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.0 | GO:0090244 | planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.0 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:2000851 | positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.0 | 0.1 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:0033986 | response to methanol(GO:0033986) |
| 0.0 | 0.0 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.0 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.0 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.1 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.0 | GO:1905073 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.0 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.0 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.0 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.0 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.1 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.0 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.1 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 0.0 | 0.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.0 | 0.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0090534 | calcium ion-transporting ATPase complex(GO:0090534) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.2 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.0 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.0 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.0 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.0 | 0.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 1.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0005148 | 3-keto sterol reductase activity(GO:0000253) prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.0 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.0 | 0.0 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.0 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.0 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 0.0 | 0.0 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 0.0 | 0.1 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.0 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.0 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.0 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.0 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) vascular endothelial growth factor binding(GO:0038085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |