Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
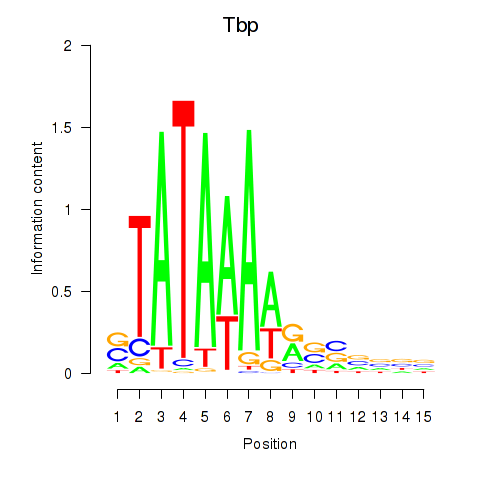
Results for Tbp
Z-value: 3.90
Transcription factors associated with Tbp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbp
|
ENSRNOG00000001489 | TATA box binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbp | rn6_v1_chr1_+_57491643_57491643 | -0.53 | 3.6e-01 | Click! |
Activity profile of Tbp motif
Sorted Z-values of Tbp motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_123106694 | 7.87 |
ENSRNOT00000028829
|
Oxt
|
oxytocin/neurophysin 1 prepropeptide |
| chr1_+_168957460 | 4.60 |
ENSRNOT00000090745
|
LOC103694857
|
hemoglobin subunit beta-2 |
| chr1_+_168945449 | 4.02 |
ENSRNOT00000087661
ENSRNOT00000019913 |
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr7_+_28654733 | 3.99 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr3_-_123119460 | 3.91 |
ENSRNOT00000028833
|
Avp
|
arginine vasopressin |
| chr1_+_168964202 | 3.34 |
ENSRNOT00000089102
|
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr3_-_82856171 | 3.32 |
ENSRNOT00000088555
|
RGD1564664
|
similar to LOC387763 protein |
| chr13_-_61070599 | 3.26 |
ENSRNOT00000005251
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr4_-_80395502 | 3.19 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chr12_-_23841049 | 3.05 |
ENSRNOT00000031555
|
Hspb1
|
heat shock protein family B (small) member 1 |
| chr1_-_168972725 | 2.56 |
ENSRNOT00000090422
|
Hbb
|
hemoglobin subunit beta |
| chrX_-_23187341 | 2.55 |
ENSRNOT00000000180
|
Alas2
|
5'-aminolevulinate synthase 2 |
| chr17_-_13393243 | 2.48 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr2_-_30127269 | 2.47 |
ENSRNOT00000023869
|
Cartpt
|
CART prepropeptide |
| chr17_-_43807540 | 2.43 |
ENSRNOT00000074763
|
LOC684762
|
similar to CG31613-PA |
| chr1_-_89369960 | 2.42 |
ENSRNOT00000028545
|
Hamp
|
hepcidin antimicrobial peptide |
| chr20_-_9855443 | 2.15 |
ENSRNOT00000090275
ENSRNOT00000066266 |
Tff3
|
trefoil factor 3 |
| chr1_+_101178104 | 1.99 |
ENSRNOT00000028072
|
Pth2
|
parathyroid hormone 2 |
| chr7_-_44121130 | 1.92 |
ENSRNOT00000005706
|
Nts
|
neurotensin |
| chr8_-_66863476 | 1.90 |
ENSRNOT00000018820
|
Rplp1
|
ribosomal protein lateral stalk subunit P1 |
| chr3_+_72385666 | 1.85 |
ENSRNOT00000011168
|
Prg2
|
proteoglycan 2 |
| chr15_-_23969011 | 1.83 |
ENSRNOT00000014821
|
Gch1
|
GTP cyclohydrolase 1 |
| chr1_-_218657925 | 1.76 |
ENSRNOT00000020425
|
Gal
|
galanin and GMAP prepropeptide |
| chr17_+_43627930 | 1.73 |
ENSRNOT00000081719
|
LOC102549061
|
histone H2B type 1-N-like |
| chr4_-_119889949 | 1.68 |
ENSRNOT00000033687
|
H1fx
|
H1 histone family, member X |
| chr19_+_41482728 | 1.67 |
ENSRNOT00000022943
|
Calb2
|
calbindin 2 |
| chr1_+_279798187 | 1.63 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr11_+_31389514 | 1.62 |
ENSRNOT00000000325
|
Olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr2_+_198388809 | 1.59 |
ENSRNOT00000083087
|
Hist2h2aa3
|
histone cluster 2, H2aa3 |
| chr18_-_15540177 | 1.56 |
ENSRNOT00000022113
|
Ttr
|
transthyretin |
| chr6_+_28382962 | 1.55 |
ENSRNOT00000016976
|
Pomc
|
proopiomelanocortin |
| chr12_+_22641104 | 1.54 |
ENSRNOT00000001916
|
Serpine1
|
serpin family E member 1 |
| chr2_-_198382190 | 1.54 |
ENSRNOT00000044268
|
Hist2h2aa2
|
histone cluster 2, H2aa2 |
| chr2_-_198360678 | 1.52 |
ENSRNOT00000051917
|
Hist2h2ac
|
histone cluster 2 H2A family member c |
| chr2_-_190100276 | 1.51 |
ENSRNOT00000015351
|
S100a9
|
S100 calcium binding protein A9 |
| chr20_+_8484311 | 1.50 |
ENSRNOT00000050041
|
LOC100364116
|
ribosomal protein S20-like |
| chr7_-_54855557 | 1.50 |
ENSRNOT00000039475
|
Glipr1l1
|
GLI pathogenesis-related 1 like 1 |
| chr2_+_198390166 | 1.47 |
ENSRNOT00000081042
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr17_-_44793927 | 1.47 |
ENSRNOT00000086309
|
Hist1h2bo
|
histone cluster 1 H2B family member o |
| chr3_-_105512939 | 1.46 |
ENSRNOT00000011773
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr13_-_89565813 | 1.43 |
ENSRNOT00000004347
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr10_+_82292110 | 1.42 |
ENSRNOT00000004435
|
Chad
|
chondroadherin |
| chr17_+_44794130 | 1.42 |
ENSRNOT00000077571
|
Hist1h2ac
|
histone cluster 1, H2ac |
| chr16_+_20352480 | 1.38 |
ENSRNOT00000025956
|
Arrdc2
|
arrestin domain containing 2 |
| chr5_+_167952728 | 1.35 |
ENSRNOT00000085315
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr19_-_26094756 | 1.28 |
ENSRNOT00000067780
|
Junb
|
JunB proto-oncogene, AP-1 transcription factor subunit |
| chr3_-_48451650 | 1.26 |
ENSRNOT00000007356
|
Gcg
|
glucagon |
| chr7_-_29171783 | 1.25 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr2_-_198380836 | 1.23 |
ENSRNOT00000040906
|
LOC102548682
|
histone H4-like |
| chr14_-_18733391 | 1.23 |
ENSRNOT00000003745
|
Cxcl2
|
C-X-C motif chemokine ligand 2 |
| chr7_-_18683440 | 1.21 |
ENSRNOT00000068323
|
Rps28
|
ribosomal protein S28 |
| chr4_-_98342887 | 1.18 |
ENSRNOT00000010156
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr4_-_181477281 | 1.16 |
ENSRNOT00000055463
|
Mansc4
|
MANSC domain containing 4 |
| chr10_+_110138586 | 1.12 |
ENSRNOT00000086096
|
Slc16a3
|
solute carrier family 16 member 3 |
| chr10_-_88172910 | 1.11 |
ENSRNOT00000046956
|
Krt42
|
keratin 42 |
| chr8_+_73593310 | 1.11 |
ENSRNOT00000012048
|
C2cd4b
|
C2 calcium-dependent domain containing 4B |
| chr17_-_43689311 | 1.10 |
ENSRNOT00000028779
|
Hist1h2bcl1
|
histone cluster 1, H2bc-like 1 |
| chr7_+_18683553 | 1.08 |
ENSRNOT00000009425
|
Ndufa7
|
NADH:ubiquinone oxidoreductase subunit A7 |
| chr10_+_71217966 | 1.07 |
ENSRNOT00000076192
|
Hnf1b
|
HNF1 homeobox B |
| chr1_-_25839198 | 1.06 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr1_+_213595240 | 1.04 |
ENSRNOT00000017137
|
Scgb1c1
|
secretoglobin, family 1C, member 1 |
| chr3_+_148215540 | 1.03 |
ENSRNOT00000029660
|
Id1
|
inhibitor of DNA binding 1, HLH protein |
| chr2_+_198417619 | 1.02 |
ENSRNOT00000085945
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr5_-_16706909 | 1.00 |
ENSRNOT00000011314
|
Rps20
|
ribosomal protein S20 |
| chr6_+_26602144 | 0.97 |
ENSRNOT00000008037
|
Ucn
|
urocortin |
| chr17_+_44522140 | 0.97 |
ENSRNOT00000080490
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr13_-_73390393 | 0.94 |
ENSRNOT00000067253
ENSRNOT00000093438 |
Lhx4
|
LIM homeobox 4 |
| chr19_-_57333433 | 0.93 |
ENSRNOT00000024917
|
Agt
|
angiotensinogen |
| chr19_-_25166528 | 0.91 |
ENSRNOT00000007775
|
Rln3
|
relaxin 3 |
| chr3_+_5709236 | 0.90 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr2_+_115362799 | 0.89 |
ENSRNOT00000015756
|
Rpl22l1
|
ribosomal protein L22 like 1 |
| chr1_-_214252456 | 0.88 |
ENSRNOT00000023504
|
Irf7
|
interferon regulatory factor 7 |
| chr7_-_143453544 | 0.88 |
ENSRNOT00000034450
ENSRNOT00000083956 |
Krt1
Krt5
|
keratin 1 keratin 5 |
| chr7_-_12882753 | 0.87 |
ENSRNOT00000011275
ENSRNOT00000044275 |
Bsg
|
basigin |
| chr17_-_43675934 | 0.86 |
ENSRNOT00000081345
|
Hist1h1t
|
histone cluster 1 H1 family member t |
| chr1_+_101412736 | 0.86 |
ENSRNOT00000067468
|
Lhb
|
luteinizing hormone beta polypeptide |
| chr2_-_251532312 | 0.86 |
ENSRNOT00000019501
|
Cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr19_-_25961666 | 0.84 |
ENSRNOT00000004091
|
Calr
|
calreticulin |
| chr9_-_73948583 | 0.81 |
ENSRNOT00000018097
|
Myl1
|
myosin, light chain 1 |
| chr19_-_11308740 | 0.80 |
ENSRNOT00000067391
|
Mt2A
|
metallothionein 2A |
| chrX_-_139916883 | 0.80 |
ENSRNOT00000090442
|
Gpc3
|
glypican 3 |
| chr20_+_5441876 | 0.80 |
ENSRNOT00000092476
|
Rps18
|
ribosomal protein S18 |
| chr1_+_72580424 | 0.78 |
ENSRNOT00000022977
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr10_+_14088319 | 0.78 |
ENSRNOT00000019508
|
Rps2
|
ribosomal protein S2 |
| chrX_-_156440461 | 0.77 |
ENSRNOT00000083951
|
Rpl10
|
ribosomal protein L10 |
| chr17_-_44744902 | 0.77 |
ENSRNOT00000085381
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr10_+_88490798 | 0.77 |
ENSRNOT00000023872
|
Cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr7_+_117519075 | 0.75 |
ENSRNOT00000029768
|
Scx
|
scleraxis bHLH transcription factor |
| chr2_-_19808937 | 0.74 |
ENSRNOT00000044237
|
Atp6ap1l
|
ATPase H+ transporting accessory protein 1 like |
| chr10_-_104575890 | 0.74 |
ENSRNOT00000050223
|
H3f3b
|
H3 histone family member 3B |
| chr4_-_145678066 | 0.74 |
ENSRNOT00000014103
|
Ghrl
|
ghrelin and obestatin prepropeptide |
| chr5_+_168078748 | 0.73 |
ENSRNOT00000024798
|
Uts2
|
urotensin 2 |
| chr3_+_149643481 | 0.72 |
ENSRNOT00000020713
|
Bpifa5
|
BPI fold containing family A, member 5 |
| chr8_+_99568958 | 0.71 |
ENSRNOT00000073400
|
Plscr5
|
phospholipid scramblase family, member 5 |
| chr10_+_72272248 | 0.69 |
ENSRNOT00000003908
|
Car4
|
carbonic anhydrase 4 |
| chr2_-_188413219 | 0.69 |
ENSRNOT00000065065
|
Fdps
|
farnesyl diphosphate synthase |
| chr10_-_15610826 | 0.67 |
ENSRNOT00000027851
|
Hbz
|
hemoglobin subunit zeta |
| chr5_+_79179417 | 0.67 |
ENSRNOT00000010454
|
Orm1
|
orosomucoid 1 |
| chr17_-_43798383 | 0.66 |
ENSRNOT00000075069
|
LOC684828
|
similar to Histone H1.2 (H1 VAR.1) (H1c) |
| chr3_+_159569363 | 0.65 |
ENSRNOT00000064159
|
Tox2
|
TOX high mobility group box family member 2 |
| chr17_+_31441630 | 0.65 |
ENSRNOT00000083705
ENSRNOT00000023582 |
Tubb2b
|
tubulin, beta 2B class IIb |
| chr1_+_256370850 | 0.65 |
ENSRNOT00000030962
|
Cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1 |
| chr10_-_87232723 | 0.63 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr1_+_91363492 | 0.62 |
ENSRNOT00000014517
|
Cebpa
|
CCAAT/enhancer binding protein alpha |
| chr3_+_160852164 | 0.62 |
ENSRNOT00000019127
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr4_+_45000973 | 0.61 |
ENSRNOT00000084736
ENSRNOT00000080880 |
ST7
|
suppression of tumorigenicity 7 |
| chr4_-_100883275 | 0.60 |
ENSRNOT00000022846
|
LOC100364435
|
thymosin, beta 10-like |
| chr8_-_56393233 | 0.59 |
ENSRNOT00000016263
|
Fdx1
|
ferredoxin 1 |
| chr6_-_104290579 | 0.59 |
ENSRNOT00000066014
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr10_-_70744315 | 0.58 |
ENSRNOT00000014865
|
Ccl5
|
C-C motif chemokine ligand 5 |
| chr4_+_168832910 | 0.58 |
ENSRNOT00000011134
|
Gprc5a
|
G protein-coupled receptor, class C, group 5, member A |
| chr2_+_208750356 | 0.57 |
ENSRNOT00000041562
|
Chia
|
chitinase, acidic |
| chr15_-_34612432 | 0.57 |
ENSRNOT00000090206
|
Mcpt1l1
|
mast cell protease 1-like 1 |
| chr11_-_32550539 | 0.57 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chr5_+_145079803 | 0.57 |
ENSRNOT00000084202
|
Sfpq
|
splicing factor proline and glutamine rich |
| chr2_-_205212681 | 0.56 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chr6_+_109300433 | 0.56 |
ENSRNOT00000010712
|
Fos
|
FBJ osteosarcoma oncogene |
| chr20_+_10438444 | 0.56 |
ENSRNOT00000071248
ENSRNOT00000075545 |
Cryaa
|
crystallin, alpha A |
| chr1_-_164441167 | 0.56 |
ENSRNOT00000023935
|
Rps3
|
ribosomal protein S3 |
| chr6_-_135112775 | 0.56 |
ENSRNOT00000086310
|
LOC103692716
|
heat shock protein HSP 90-alpha |
| chr17_+_31493107 | 0.56 |
ENSRNOT00000023611
ENSRNOT00000086264 |
Tubb2a
|
tubulin, beta 2A class IIa |
| chr9_-_37231291 | 0.55 |
ENSRNOT00000016237
|
Ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr18_+_61563053 | 0.54 |
ENSRNOT00000022845
|
Grp
|
gastrin releasing peptide |
| chr2_-_45518502 | 0.54 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chrX_-_121731543 | 0.53 |
ENSRNOT00000018788
|
Klhl13
|
kelch-like family member 13 |
| chr10_-_88163712 | 0.52 |
ENSRNOT00000005382
ENSRNOT00000084493 |
Krt17
|
keratin 17 |
| chr8_+_75687100 | 0.52 |
ENSRNOT00000038677
|
Anxa2
|
annexin A2 |
| chr7_-_11754508 | 0.51 |
ENSRNOT00000026341
|
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr1_-_224921092 | 0.51 |
ENSRNOT00000025196
|
Slc3a2
|
solute carrier family 3 member 2 |
| chr1_+_79894450 | 0.51 |
ENSRNOT00000057963
|
Nanos2
|
nanos C2HC-type zinc finger 2 |
| chr10_+_109774878 | 0.51 |
ENSRNOT00000054954
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr11_+_47243342 | 0.50 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr2_+_208900103 | 0.50 |
ENSRNOT00000055966
|
Chi3l3
|
chitinase 3-like 3 |
| chr8_+_50559126 | 0.50 |
ENSRNOT00000024918
|
Apoa5
|
apolipoprotein A5 |
| chr9_-_97151832 | 0.50 |
ENSRNOT00000040169
|
Asb18
|
ankyrin repeat and SOCS box-containing 18 |
| chr7_-_118108864 | 0.50 |
ENSRNOT00000006184
|
Mb
|
myoglobin |
| chr16_+_20121352 | 0.50 |
ENSRNOT00000025347
|
Insl3
|
insulin-like 3 |
| chr7_-_121302740 | 0.50 |
ENSRNOT00000067032
|
Rpl3
|
ribosomal protein L3 |
| chr3_+_149221377 | 0.49 |
ENSRNOT00000016220
ENSRNOT00000087752 |
Mapre1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr3_-_111422203 | 0.49 |
ENSRNOT00000084290
|
Oip5
|
Opa interacting protein 5 |
| chr2_+_208749996 | 0.49 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr1_+_89008117 | 0.47 |
ENSRNOT00000028401
|
Hspb6
|
heat shock protein family B (small) member 6 |
| chr13_-_88497901 | 0.47 |
ENSRNOT00000058560
|
Uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr14_-_85191557 | 0.47 |
ENSRNOT00000011604
|
Nefh
|
neurofilament heavy |
| chr2_+_93792601 | 0.47 |
ENSRNOT00000014701
ENSRNOT00000077311 |
Fabp4
|
fatty acid binding protein 4 |
| chr10_-_88036040 | 0.47 |
ENSRNOT00000018851
|
Krt13
|
keratin 13 |
| chr7_-_143497108 | 0.46 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr16_+_49266903 | 0.46 |
ENSRNOT00000014704
|
Slc25a4
|
solute carrier family 25 member 4 |
| chr20_+_33077106 | 0.46 |
ENSRNOT00000000456
|
Vgll2
|
vestigial-like family member 2 |
| chr5_+_24434872 | 0.46 |
ENSRNOT00000010696
|
Ccne2
|
cyclin E2 |
| chr7_-_12810570 | 0.45 |
ENSRNOT00000012578
|
Fstl3
|
follistatin like 3 |
| chr4_-_101156576 | 0.44 |
ENSRNOT00000020894
|
Dnah6
|
dynein, axonemal, heavy chain 6 |
| chr1_-_153752541 | 0.44 |
ENSRNOT00000080927
|
Prss23
|
protease, serine, 23 |
| chr15_+_44441856 | 0.44 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr17_-_44758170 | 0.44 |
ENSRNOT00000091176
|
Hist1h2bo
|
histone cluster 1 H2B family member o |
| chr10_-_91122018 | 0.44 |
ENSRNOT00000080077
|
Dcakd
|
dephospho-CoA kinase domain containing |
| chr17_-_43821536 | 0.44 |
ENSRNOT00000072286
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chrX_+_83926513 | 0.44 |
ENSRNOT00000035274
|
RGD1561958
|
similar to RIKEN cDNA 2010106E10 |
| chr1_-_65664767 | 0.44 |
ENSRNOT00000026528
ENSRNOT00000088367 |
Rps5
|
ribosomal protein S5 |
| chr7_-_145338152 | 0.43 |
ENSRNOT00000055268
|
Spt1
|
salivary protein 1 |
| chr19_-_11057254 | 0.43 |
ENSRNOT00000025559
|
Herpud1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr1_+_226030938 | 0.43 |
ENSRNOT00000030919
|
Fth1
|
ferritin heavy chain 1 |
| chr10_+_31532250 | 0.43 |
ENSRNOT00000009238
|
Med7
|
mediator complex subunit 7 |
| chr9_+_17817721 | 0.43 |
ENSRNOT00000086986
ENSRNOT00000026920 |
Hsp90ab1
|
heat shock protein 90 alpha family class B member 1 |
| chr20_+_28920616 | 0.43 |
ENSRNOT00000070897
|
P4ha1
|
prolyl 4-hydroxylase subunit alpha 1 |
| chr2_+_189997129 | 0.43 |
ENSRNOT00000015958
|
S100a4
|
S100 calcium-binding protein A4 |
| chr17_+_80882666 | 0.42 |
ENSRNOT00000024430
|
Vim
|
vimentin |
| chr4_+_79573998 | 0.42 |
ENSRNOT00000074351
|
LOC100912228
|
pro-neuropeptide Y-like |
| chr8_+_48665652 | 0.42 |
ENSRNOT00000059715
|
H2afx
|
H2A histone family, member X |
| chr9_+_69953440 | 0.42 |
ENSRNOT00000034740
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr5_-_159336429 | 0.41 |
ENSRNOT00000009230
|
Padi3
|
peptidyl arginine deiminase 3 |
| chr1_+_282134981 | 0.41 |
ENSRNOT00000036203
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr5_+_148528725 | 0.41 |
ENSRNOT00000017325
|
Fabp3
|
fatty acid binding protein 3 |
| chr10_-_75120247 | 0.40 |
ENSRNOT00000011402
|
Lpo
|
lactoperoxidase |
| chr4_-_116916236 | 0.40 |
ENSRNOT00000020749
|
Spr
|
sepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase) |
| chr9_+_46840992 | 0.40 |
ENSRNOT00000019415
|
Il1r2
|
interleukin 1 receptor type 2 |
| chr1_+_216237697 | 0.40 |
ENSRNOT00000027760
|
Cd81
|
Cd81 molecule |
| chr1_-_85220237 | 0.40 |
ENSRNOT00000026907
|
Sycn
|
syncollin |
| chr8_-_13251812 | 0.40 |
ENSRNOT00000049724
ENSRNOT00000078117 |
Piwil4
|
piwi-like RNA-mediated gene silencing 4 |
| chr4_-_170860225 | 0.40 |
ENSRNOT00000007577
|
Mgp
|
matrix Gla protein |
| chr4_+_79557854 | 0.40 |
ENSRNOT00000013145
|
Npy
|
neuropeptide Y |
| chr11_-_25350974 | 0.39 |
ENSRNOT00000002187
|
Adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr1_-_100537377 | 0.39 |
ENSRNOT00000026599
|
Spib
|
Spi-B transcription factor |
| chr12_+_23473270 | 0.39 |
ENSRNOT00000001935
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr7_-_140640953 | 0.39 |
ENSRNOT00000083156
|
Tuba1a
|
tubulin, alpha 1A |
| chr20_-_11556312 | 0.39 |
ENSRNOT00000091856
|
RGD1561557
|
similar to chromosome 21 open reading frame 29 |
| chr10_-_104075777 | 0.39 |
ENSRNOT00000004862
|
Hn1
|
hematological and neurological expressed 1 |
| chr3_+_114087287 | 0.39 |
ENSRNOT00000023017
|
B2m
|
beta-2 microglobulin |
| chr16_+_20555395 | 0.38 |
ENSRNOT00000026652
|
Gdf15
|
growth differentiation factor 15 |
| chr7_+_121480723 | 0.37 |
ENSRNOT00000065304
|
Atf4
|
activating transcription factor 4 |
| chr17_+_23116661 | 0.37 |
ENSRNOT00000067374
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr7_+_37812831 | 0.37 |
ENSRNOT00000005910
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr11_+_30363280 | 0.37 |
ENSRNOT00000002885
|
Sod1
|
superoxide dismutase 1, soluble |
| chr19_-_26084679 | 0.37 |
ENSRNOT00000046181
|
Rnaseh2a
|
ribonuclease H2, subunit A |
| chr1_+_80321585 | 0.37 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr15_+_44799334 | 0.37 |
ENSRNOT00000018599
|
Nefl
|
neurofilament light |
| chr11_-_81379640 | 0.37 |
ENSRNOT00000002484
|
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr6_+_3657325 | 0.36 |
ENSRNOT00000010927
|
Tmem178a
|
transmembrane protein 178A |
| chr14_+_11256268 | 0.36 |
ENSRNOT00000047840
ENSRNOT00000003173 ENSRNOT00000003158 |
Hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr2_+_211930326 | 0.36 |
ENSRNOT00000027765
|
Slc25a24
|
solute carrier family 25 member 24 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbp
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.6 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 1.4 | 15.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.0 | 3.0 | GO:1903544 | cellular response to interleukin-11(GO:0071348) response to butyrate(GO:1903544) |
| 0.8 | 2.5 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.8 | 4.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.6 | 2.5 | GO:1901423 | response to benzene(GO:1901423) |
| 0.6 | 2.5 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.6 | 1.8 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.6 | 1.8 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.6 | 1.8 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.6 | 2.4 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) cellular response to bile acid(GO:1903413) |
| 0.5 | 3.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.5 | 1.6 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.5 | 1.5 | GO:1990823 | response to resveratrol(GO:1904638) response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.5 | 1.0 | GO:0035482 | gastric motility(GO:0035482) |
| 0.5 | 0.5 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.5 | 2.3 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 0.4 | 1.3 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.4 | 0.8 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.4 | 1.9 | GO:1904401 | cellular response to Thyroid stimulating hormone(GO:1904401) |
| 0.4 | 1.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.4 | 0.4 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) |
| 0.4 | 1.1 | GO:0061017 | regulation of pronephros size(GO:0035565) pronephric nephron tubule development(GO:0039020) hepatoblast differentiation(GO:0061017) |
| 0.4 | 1.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.3 | 0.9 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.3 | 3.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.3 | 0.9 | GO:0001543 | ovarian follicle rupture(GO:0001543) angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of renal output by angiotensin(GO:0002019) angiotensin-mediated drinking behavior(GO:0003051) regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) positive regulation of gap junction assembly(GO:1903598) |
| 0.3 | 1.2 | GO:1903576 | response to L-arginine(GO:1903576) |
| 0.3 | 0.8 | GO:0002397 | MHC class I protein complex assembly(GO:0002397) |
| 0.3 | 0.8 | GO:1903937 | neurofilament bundle assembly(GO:0033693) response to acrylamide(GO:1903937) |
| 0.3 | 0.8 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.3 | 1.5 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.3 | 1.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.3 | 0.5 | GO:0015827 | tryptophan transport(GO:0015827) |
| 0.3 | 0.8 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.2 | 2.4 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.2 | 0.7 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.2 | 1.2 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.2 | 5.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 1.0 | GO:0061741 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 0.2 | 0.8 | GO:0035519 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 0.6 | GO:0042531 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) diapedesis(GO:0050904) |
| 0.2 | 0.4 | GO:0034756 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 0.2 | 0.4 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.2 | 1.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 0.5 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.2 | 0.9 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.2 | 0.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 0.6 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.2 | 1.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 0.8 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.7 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.1 | 1.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.9 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 0.4 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 2.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 0.7 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 1.4 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.1 | 0.7 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) |
| 0.1 | 0.4 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.4 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.1 | 9.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.4 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.3 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.4 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.4 | GO:1904350 | regulation of protein catabolic process in the vacuole(GO:1904350) |
| 0.1 | 0.7 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.4 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.1 | 0.5 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 0.9 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.1 | 0.6 | GO:0002892 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 1.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.3 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 0.3 | GO:2000525 | positive regulation of hypersensitivity(GO:0002885) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.6 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.1 | 0.6 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.3 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.8 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.3 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.4 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.3 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.1 | 0.3 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.1 | 0.2 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.4 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 0.5 | GO:1904528 | positive regulation of microtubule binding(GO:1904528) |
| 0.1 | 0.4 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.1 | 0.7 | GO:0015866 | ADP transport(GO:0015866) |
| 0.1 | 0.3 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.1 | 0.5 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 0.3 | GO:0019541 | propionate metabolic process(GO:0019541) negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
| 0.1 | 0.3 | GO:0046495 | nicotinamide riboside catabolic process(GO:0006738) urate biosynthetic process(GO:0034418) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.1 | 0.4 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.4 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.3 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.5 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.3 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 0.2 | GO:1990036 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.1 | 0.2 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.1 | 0.4 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 5.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 0.6 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.6 | GO:0000050 | urea cycle(GO:0000050) |
| 0.1 | 0.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 2.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.4 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 0.3 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.1 | 0.3 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.1 | 1.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.6 | GO:2000018 | regulation of male gonad development(GO:2000018) |
| 0.1 | 0.2 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.1 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.3 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 1.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.3 | GO:0044206 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) |
| 0.0 | 0.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.6 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.2 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.2 | GO:2000790 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.0 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.2 | GO:1902739 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.0 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.2 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 1.1 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.2 | GO:0015669 | gas transport(GO:0015669) |
| 0.0 | 0.6 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 1.3 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.5 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.5 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.2 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.9 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:2000458 | astrocyte chemotaxis(GO:0035700) T follicular helper cell differentiation(GO:0061470) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.0 | 0.2 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.0 | 0.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.2 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.4 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.5 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.4 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0098953 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) gamma-aminobutyric acid receptor clustering(GO:0097112) receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.1 | GO:1903209 | positive regulation of oxidative stress-induced cell death(GO:1903209) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.1 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 2.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.2 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.0 | 0.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.7 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 2.2 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.2 | GO:0044146 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.3 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.6 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.4 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.8 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.3 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.4 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.6 | GO:0034698 | response to gonadotropin(GO:0034698) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.3 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 15.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 3.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 1.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.4 | 1.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.3 | 2.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 0.6 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.3 | 0.9 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.3 | 0.8 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.3 | 22.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 0.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 0.7 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.2 | 1.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 1.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.4 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.1 | 0.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.6 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 6.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.3 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.4 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.2 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 0.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 10.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.9 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 4.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 13.5 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.4 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.4 | GO:0005767 | secondary lysosome(GO:0005767) autolysosome(GO:0044754) |
| 0.0 | 0.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 4.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 2.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 2.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.0 | 0.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 19.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 15.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.3 | 3.9 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.8 | 2.5 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.7 | 0.7 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.6 | 15.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 1.5 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.3 | 1.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.3 | 1.8 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.3 | 1.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 0.9 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.2 | 0.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 1.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 4.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 0.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 1.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.2 | 0.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.4 | GO:0032551 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 9.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 1.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.7 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 1.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.6 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.4 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 0.6 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.3 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 0.1 | 0.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.3 | GO:0042979 | ornithine decarboxylase activator activity(GO:0042978) ornithine decarboxylase regulator activity(GO:0042979) |
| 0.1 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 3.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.6 | GO:0048019 | CCR1 chemokine receptor binding(GO:0031726) receptor antagonist activity(GO:0048019) |
| 0.1 | 0.3 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 0.1 | 0.8 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.3 | GO:0004731 | purine nucleobase binding(GO:0002060) purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.4 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 2.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 3.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.8 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 0.6 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.3 | GO:0015168 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.5 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 1.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.5 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.2 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 1.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.2 | GO:0099567 | N-terminal myristoylation domain binding(GO:0031997) calcium ion binding involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099567) |
| 0.1 | 0.4 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 8.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 4.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 9.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.1 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 1.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 1.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 8.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.4 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 2.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.1 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 1.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0044325 | ion channel binding(GO:0044325) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 7.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 2.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 3.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 4.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 3.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 7.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 3.0 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.2 | 2.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 2.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 23.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 2.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 5.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 0.6 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 3.4 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 0.7 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 3.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 3.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.9 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.4 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |