Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Tal1
Z-value: 0.31
Transcription factors associated with Tal1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tal1
|
ENSRNOG00000025051 | TAL bHLH transcription factor 1, erythroid differentiation factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tal1 | rn6_v1_chr5_+_133864798_133864798 | 0.23 | 7.1e-01 | Click! |
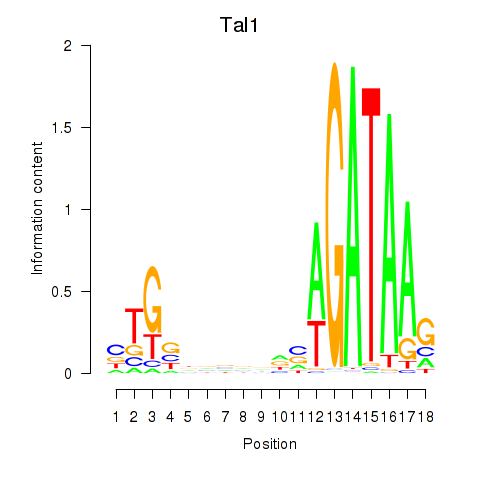
Activity profile of Tal1 motif
Sorted Z-values of Tal1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_23187341 | 0.19 |
ENSRNOT00000000180
|
Alas2
|
5'-aminolevulinate synthase 2 |
| chr10_-_15590220 | 0.18 |
ENSRNOT00000048977
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr5_+_153197459 | 0.17 |
ENSRNOT00000023187
|
Rhd
|
Rh blood group, D antigen |
| chr20_+_4593389 | 0.12 |
ENSRNOT00000001174
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr10_-_15603649 | 0.12 |
ENSRNOT00000051483
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr1_+_279798187 | 0.11 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr1_+_175445088 | 0.11 |
ENSRNOT00000036718
|
Adm
|
adrenomedullin |
| chr7_-_145174771 | 0.10 |
ENSRNOT00000055270
|
Glycam1
|
glycosylation dependent cell adhesion molecule 1 |
| chr17_-_22143324 | 0.09 |
ENSRNOT00000019361
|
Edn1
|
endothelin 1 |
| chr7_+_121841855 | 0.09 |
ENSRNOT00000024673
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr1_-_164205222 | 0.09 |
ENSRNOT00000035774
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr7_-_115981731 | 0.08 |
ENSRNOT00000036549
|
RGD1308195
|
similar to secreted Ly6/uPAR related protein 2 |
| chr7_+_138707426 | 0.07 |
ENSRNOT00000037874
|
Pced1b
|
PC-esterase domain containing 1B |
| chrX_-_121731543 | 0.07 |
ENSRNOT00000018788
|
Klhl13
|
kelch-like family member 13 |
| chr3_-_123702732 | 0.07 |
ENSRNOT00000028859
|
RGD1311739
|
similar to RIKEN cDNA 1700037H04 |
| chr10_-_16788126 | 0.07 |
ENSRNOT00000077386
|
Atp6v0e1
|
ATPase H+ transporting V0 subunit e1 |
| chr16_-_75648538 | 0.06 |
ENSRNOT00000048414
|
Defb14
|
defensin beta 14 |
| chr20_-_5806097 | 0.06 |
ENSRNOT00000000611
|
Clps
|
colipase |
| chr11_-_81344488 | 0.06 |
ENSRNOT00000002492
|
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr4_+_57855416 | 0.06 |
ENSRNOT00000029608
|
Cpa2
|
carboxypeptidase A2 |
| chr16_-_45929 | 0.06 |
ENSRNOT00000043153
|
AABR07024439.1
|
|
| chr13_-_80745347 | 0.06 |
ENSRNOT00000041908
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr14_-_84189266 | 0.05 |
ENSRNOT00000005934
|
Tcn2
|
transcobalamin 2 |
| chr2_+_208541361 | 0.05 |
ENSRNOT00000021288
|
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chr6_-_137084739 | 0.05 |
ENSRNOT00000060943
|
Tmem179
|
transmembrane protein 179 |
| chr4_-_166617143 | 0.05 |
ENSRNOT00000038077
|
AABR07062268.1
|
|
| chr15_-_45524582 | 0.05 |
ENSRNOT00000081912
|
Gucy1b2
|
guanylate cyclase 1 soluble subunit beta 2 |
| chr14_-_18704059 | 0.05 |
ENSRNOT00000081455
|
Mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr1_+_73040901 | 0.05 |
ENSRNOT00000071927
|
Gp6
|
glycoprotein 6 (platelet) |
| chr1_-_37990007 | 0.05 |
ENSRNOT00000081054
|
AABR07001100.1
|
|
| chr7_-_71269869 | 0.05 |
ENSRNOT00000037030
|
Uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr8_-_119326938 | 0.05 |
ENSRNOT00000044467
|
Ccrl2
|
C-C motif chemokine receptor like 2 |
| chr5_+_126862987 | 0.05 |
ENSRNOT00000080701
|
Hspb11
|
heat shock protein family B (small), member 11 |
| chr17_-_21677477 | 0.04 |
ENSRNOT00000035448
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr9_+_81535483 | 0.04 |
ENSRNOT00000077285
|
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr8_-_33661049 | 0.04 |
ENSRNOT00000068037
|
Fli1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr18_-_81682206 | 0.04 |
ENSRNOT00000058219
|
LOC100359752
|
hypothetical protein LOC100359752 |
| chr5_+_133864798 | 0.04 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr10_-_87407634 | 0.04 |
ENSRNOT00000016657
|
Krt23
|
keratin 23 |
| chr2_-_123851854 | 0.04 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr16_-_21338771 | 0.04 |
ENSRNOT00000014265
|
Pbx4
|
PBX homeobox 4 |
| chr13_+_83972212 | 0.04 |
ENSRNOT00000004394
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr10_-_88107232 | 0.04 |
ENSRNOT00000019330
ENSRNOT00000076770 |
Krt9
|
keratin 9 |
| chr10_-_15610826 | 0.04 |
ENSRNOT00000027851
|
Hbz
|
hemoglobin subunit zeta |
| chr7_-_33793565 | 0.04 |
ENSRNOT00000065354
|
AABR07056633.1
|
|
| chr10_-_102227554 | 0.04 |
ENSRNOT00000078035
|
AC096468.1
|
|
| chr14_+_22517774 | 0.04 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr1_-_98570949 | 0.04 |
ENSRNOT00000033648
|
Siglec5
|
sialic acid binding Ig-like lectin 5 |
| chr19_+_14835822 | 0.04 |
ENSRNOT00000072804
|
1700007B14Rik
|
RIKEN cDNA 1700007B14 gene |
| chr1_+_279896973 | 0.04 |
ENSRNOT00000068119
|
Pnliprp2
|
pancreatic lipase related protein 2 |
| chr9_-_23493081 | 0.04 |
ENSRNOT00000072144
|
Rhag
|
Rh-associated glycoprotein |
| chr7_+_60015998 | 0.04 |
ENSRNOT00000007309
|
Best3
|
bestrophin 3 |
| chr6_+_27975849 | 0.04 |
ENSRNOT00000060810
|
Dtnb
|
dystrobrevin, beta |
| chr10_-_34961608 | 0.04 |
ENSRNOT00000033056
|
LOC103689931
|
heterogeneous nuclear ribonucleoprotein A/B |
| chrX_-_63803915 | 0.04 |
ENSRNOT00000076938
|
Maged1
|
MAGE family member D1 |
| chr20_-_2191640 | 0.03 |
ENSRNOT00000001016
|
Trim10
|
tripartite motif-containing 10 |
| chr2_+_190073815 | 0.03 |
ENSRNOT00000015473
|
S100a8
|
S100 calcium binding protein A8 |
| chr5_+_164808323 | 0.03 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr17_+_49714294 | 0.03 |
ENSRNOT00000018190
|
Rala
|
RAS like proto-oncogene A |
| chr20_+_13498926 | 0.03 |
ENSRNOT00000070992
ENSRNOT00000045375 |
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr8_+_22047697 | 0.03 |
ENSRNOT00000067741
|
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr2_-_187909394 | 0.03 |
ENSRNOT00000032355
|
Rab25
|
RAB25, member RAS oncogene family |
| chr1_+_189359853 | 0.03 |
ENSRNOT00000055083
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr1_+_100501676 | 0.03 |
ENSRNOT00000043724
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr16_-_68968248 | 0.03 |
ENSRNOT00000016885
|
Eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr3_+_103945329 | 0.03 |
ENSRNOT00000008364
|
Emc7
|
ER membrane protein complex subunit 7 |
| chr3_+_5519990 | 0.03 |
ENSRNOT00000070873
ENSRNOT00000007640 |
Adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr2_-_88660449 | 0.03 |
ENSRNOT00000051741
|
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr19_-_53477420 | 0.03 |
ENSRNOT00000071344
|
LOC687560
|
hypothetical protein LOC687560 |
| chr7_-_8060373 | 0.03 |
ENSRNOT00000071699
|
Olfr798
|
olfactory receptor 798 |
| chr5_+_57845819 | 0.03 |
ENSRNOT00000017712
|
Nudt2
|
nudix hydrolase 2 |
| chr3_+_119561290 | 0.03 |
ENSRNOT00000015843
|
Blvra
|
biliverdin reductase A |
| chr4_-_119148358 | 0.03 |
ENSRNOT00000012218
|
Gkn1
|
gastrokine 1 |
| chr1_-_213635546 | 0.03 |
ENSRNOT00000018861
|
Sirt3
|
sirtuin 3 |
| chr10_-_68926264 | 0.03 |
ENSRNOT00000008661
|
Phb-ps1
|
prohibitin, pseudogene 1 |
| chr2_-_193335002 | 0.03 |
ENSRNOT00000012691
|
Lce1m
|
late cornified envelope 1M |
| chr1_+_220416018 | 0.03 |
ENSRNOT00000027233
|
B4gat1
|
beta-1,4-glucuronyltransferase 1 |
| chr12_-_38116919 | 0.03 |
ENSRNOT00000001467
|
Denr
|
density regulated reinitiation and release factor |
| chr8_+_64088913 | 0.03 |
ENSRNOT00000013022
|
Adpgk
|
ADP-dependent glucokinase |
| chr7_-_143738237 | 0.03 |
ENSRNOT00000055320
|
Spryd3
|
SPRY domain containing 3 |
| chr16_-_18645941 | 0.03 |
ENSRNOT00000079241
|
Dydc2
|
DPY30 domain containing 2 |
| chr5_+_48303366 | 0.03 |
ENSRNOT00000009973
|
Gabrr2
|
gamma-aminobutyric acid type A receptor rho 2 subunit |
| chr1_+_142883040 | 0.03 |
ENSRNOT00000015898
|
Alpk3
|
alpha-kinase 3 |
| chr8_-_63291966 | 0.03 |
ENSRNOT00000077762
|
AABR07070278.1
|
|
| chr16_-_39970532 | 0.03 |
ENSRNOT00000071331
|
Spata4
|
spermatogenesis associated 4 |
| chr4_+_7662019 | 0.03 |
ENSRNOT00000014023
|
Fam126a
|
family with sequence similarity 126, member A |
| chr14_-_110883784 | 0.03 |
ENSRNOT00000086299
ENSRNOT00000068190 |
Vrk2
|
vaccinia related kinase 2 |
| chr6_+_58388333 | 0.03 |
ENSRNOT00000036337
|
LOC680646
|
similar to 40S ribosomal protein S2 |
| chr10_+_105630810 | 0.03 |
ENSRNOT00000064904
|
Prcd
|
photoreceptor disc component |
| chr10_-_89374516 | 0.03 |
ENSRNOT00000028084
|
Vat1
|
vesicle amine transport 1 |
| chr8_-_17525906 | 0.03 |
ENSRNOT00000007855
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr18_-_37096132 | 0.02 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr5_+_144701949 | 0.02 |
ENSRNOT00000015618
|
Psmb2
|
proteasome subunit beta 2 |
| chrX_-_16537888 | 0.02 |
ENSRNOT00000060247
|
Dgkk
|
diacylglycerol kinase kappa |
| chr10_+_10967658 | 0.02 |
ENSRNOT00000004999
|
Cdip1
|
cell death-inducing p53 target 1 |
| chr10_+_83201311 | 0.02 |
ENSRNOT00000006179
|
Slc35b1
|
solute carrier family 35, member B1 |
| chr7_-_121000272 | 0.02 |
ENSRNOT00000020941
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr4_-_145300177 | 0.02 |
ENSRNOT00000029728
|
Camk1
|
calcium/calmodulin-dependent protein kinase I |
| chr8_-_66863476 | 0.02 |
ENSRNOT00000018820
|
Rplp1
|
ribosomal protein lateral stalk subunit P1 |
| chr8_-_48672732 | 0.02 |
ENSRNOT00000079275
|
Hmbs
|
hydroxymethylbilane synthase |
| chr9_-_113358526 | 0.02 |
ENSRNOT00000065073
|
Txndc2
|
thioredoxin domain containing 2 |
| chr19_-_42920344 | 0.02 |
ENSRNOT00000019408
ENSRNOT00000068594 |
Zfhx3
|
zinc finger homeobox 3 |
| chr1_+_213676954 | 0.02 |
ENSRNOT00000050551
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr10_+_53740841 | 0.02 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr16_+_51748970 | 0.02 |
ENSRNOT00000059182
|
Adam26a
|
a disintegrin and metallopeptidase domain 26A |
| chr18_+_35136132 | 0.02 |
ENSRNOT00000078731
|
Spink5
|
serine peptidase inhibitor, Kazal type 5 |
| chr4_+_29535852 | 0.02 |
ENSRNOT00000087619
|
NEWGENE_621351
|
collagen, type I, alpha 2 |
| chr15_+_59678165 | 0.02 |
ENSRNOT00000074868
|
Enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr7_-_116504853 | 0.02 |
ENSRNOT00000056557
|
RGD1565410
|
similar to Ly6-C antigen gene |
| chr1_+_88620317 | 0.02 |
ENSRNOT00000075116
|
Cox7a1
|
cytochrome c oxidase subunit 7A1 |
| chr4_-_165195489 | 0.02 |
ENSRNOT00000079429
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr1_-_100501437 | 0.02 |
ENSRNOT00000026445
|
Emc10
|
ER membrane protein complex subunit 10 |
| chr19_-_49623758 | 0.02 |
ENSRNOT00000071130
|
Pkd1l2
|
polycystic kidney disease 1-like 2 |
| chr10_-_90415070 | 0.02 |
ENSRNOT00000055179
|
Itga2b
|
integrin subunit alpha 2b |
| chr2_+_85377318 | 0.02 |
ENSRNOT00000016506
ENSRNOT00000085094 |
Sema5a
|
semaphorin 5A |
| chr1_+_81499821 | 0.02 |
ENSRNOT00000027100
|
Lypd3
|
Ly6/Plaur domain containing 3 |
| chr17_+_76008807 | 0.02 |
ENSRNOT00000070895
|
Echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr3_-_161246351 | 0.02 |
ENSRNOT00000020348
|
Tnnc2
|
troponin C2, fast skeletal type |
| chr16_-_82999827 | 0.02 |
ENSRNOT00000041223
|
Tex29
|
testis expressed 29 |
| chr1_-_153752541 | 0.02 |
ENSRNOT00000080927
|
Prss23
|
protease, serine, 23 |
| chrX_-_135342996 | 0.02 |
ENSRNOT00000084848
ENSRNOT00000008503 |
Aifm1
|
apoptosis inducing factor, mitochondria associated 1 |
| chr7_+_120153184 | 0.02 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chr6_+_102477036 | 0.02 |
ENSRNOT00000087132
|
Rad51b
|
RAD51 paralog B |
| chr5_-_78324278 | 0.02 |
ENSRNOT00000082642
ENSRNOT00000048904 |
Wdr31
|
WD repeat domain 31 |
| chr9_-_113357933 | 0.02 |
ENSRNOT00000032330
|
Txndc2
|
thioredoxin domain containing 2 |
| chr10_+_106785077 | 0.02 |
ENSRNOT00000075047
|
Tmc8
|
transmembrane channel-like 8 |
| chr2_+_212257225 | 0.02 |
ENSRNOT00000077883
|
Vav3
|
vav guanine nucleotide exchange factor 3 |
| chrX_+_13441558 | 0.02 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chr13_-_72869396 | 0.02 |
ENSRNOT00000093563
|
RGD1304622
|
similar to 6820428L09 protein |
| chr4_-_61720956 | 0.02 |
ENSRNOT00000012879
|
Akr1b1
|
aldo-keto reductase family 1 member B |
| chr5_-_133067245 | 0.02 |
ENSRNOT00000033163
|
Skint10
|
selection and upkeep of intraepithelial T cells 10 |
| chr2_+_142717281 | 0.02 |
ENSRNOT00000014765
ENSRNOT00000076774 |
Stoml3
|
stomatin like 3 |
| chr13_+_51218468 | 0.02 |
ENSRNOT00000033636
|
LOC289035
|
similar to UDP-N-acetylglucosamine:a-1,3-D-mannoside beta-1,4-N-acetylgluco |
| chr13_-_51373399 | 0.02 |
ENSRNOT00000060656
|
Mgat4e
|
MGAT4 family, member E |
| chr10_-_103919605 | 0.02 |
ENSRNOT00000004718
|
Hid1
|
HID1 domain containing |
| chr15_+_33606124 | 0.02 |
ENSRNOT00000065210
|
AC115371.1
|
|
| chr16_+_61926586 | 0.02 |
ENSRNOT00000058884
|
AABR07026059.1
|
|
| chr3_-_148706972 | 0.02 |
ENSRNOT00000036956
|
Tspy26
|
testis specific protein, Y-linked 26 |
| chr1_+_213686046 | 0.02 |
ENSRNOT00000019808
|
LOC108348167
|
NACHT, LRR and PYD domains-containing protein 6-like |
| chr1_+_260289589 | 0.02 |
ENSRNOT00000051058
|
Dntt
|
DNA nucleotidylexotransferase |
| chrX_-_135250519 | 0.02 |
ENSRNOT00000044487
|
Elf4
|
E74 like ETS transcription factor 4 |
| chr1_+_198210525 | 0.02 |
ENSRNOT00000026755
|
Ypel3
|
yippee-like 3 |
| chrX_-_84167717 | 0.02 |
ENSRNOT00000006415
|
Pof1b
|
premature ovarian failure 1B |
| chr13_+_76962504 | 0.02 |
ENSRNOT00000076581
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr10_-_37055282 | 0.02 |
ENSRNOT00000086193
ENSRNOT00000065584 |
Hnrnpab
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr12_-_24365324 | 0.02 |
ENSRNOT00000032250
|
Trim50
|
tripartite motif-containing 50 |
| chr2_+_24668116 | 0.02 |
ENSRNOT00000067022
ENSRNOT00000036748 |
Wdr41
|
WD repeat domain 41 |
| chr15_+_34446878 | 0.02 |
ENSRNOT00000027619
|
Ltb4r2
|
leukotriene B4 receptor 2 |
| chrX_+_124631881 | 0.02 |
ENSRNOT00000009329
|
Atp1b4
|
ATPase Na+/K+ transporting family member beta 4 |
| chr4_+_82776746 | 0.02 |
ENSRNOT00000011371
|
Tax1bp1
|
Tax1 binding protein 1 |
| chr12_+_17252093 | 0.02 |
ENSRNOT00000047634
|
Zfand2a
|
zinc finger AN1-type containing 2A |
| chr3_+_147511406 | 0.02 |
ENSRNOT00000082438
|
Slc52a3
|
solute carrier family 52 member 3 |
| chr16_+_14306804 | 0.02 |
ENSRNOT00000017773
|
Lrit1
|
leucine-rich repeat, Ig-like and transmembrane domains 1 |
| chr1_-_147077568 | 0.02 |
ENSRNOT00000074040
|
LOC100911002
|
synaptonemal complex protein 3-like |
| chr1_-_222459952 | 0.02 |
ENSRNOT00000028752
|
Otub1
|
OTU deubiquitinase, ubiquitin aldehyde binding 1 |
| chr1_+_86972244 | 0.02 |
ENSRNOT00000036907
|
Rinl
|
Ras and Rab interactor-like |
| chr4_-_165541314 | 0.02 |
ENSRNOT00000013833
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr9_+_44025020 | 0.02 |
ENSRNOT00000024582
|
Unc50
|
unc-50 inner nuclear membrane RNA binding protein |
| chr1_+_227640680 | 0.02 |
ENSRNOT00000033613
|
Ms4a4c
|
membrane-spanning 4-domains, subfamily A, member 4C |
| chr1_-_201110928 | 0.02 |
ENSRNOT00000027721
|
Nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr4_+_56337695 | 0.02 |
ENSRNOT00000071926
|
Lep
|
leptin |
| chr5_+_173373888 | 0.02 |
ENSRNOT00000077602
|
Ube2j2
|
ubiquitin-conjugating enzyme E2, J2 |
| chr10_+_70417108 | 0.02 |
ENSRNOT00000079325
|
Slfn4
|
schlafen 4 |
| chr14_-_78469480 | 0.02 |
ENSRNOT00000006771
|
Jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr4_+_147333056 | 0.02 |
ENSRNOT00000012137
|
Pparg
|
peroxisome proliferator-activated receptor gamma |
| chr4_+_14070553 | 0.02 |
ENSRNOT00000077505
|
Cd36
|
CD36 molecule |
| chr8_-_86286991 | 0.02 |
ENSRNOT00000046322
|
LOC367117
|
similar to RIKEN cDNA 2900055D03 |
| chr1_+_260732485 | 0.02 |
ENSRNOT00000076842
ENSRNOT00000076073 |
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr7_-_17520780 | 0.02 |
ENSRNOT00000029672
|
RGD1560291
|
similar to NACHT, leucine rich repeat and PYD containing 4A |
| chr13_+_52645257 | 0.01 |
ENSRNOT00000012801
|
Lad1
|
ladinin 1 |
| chr4_+_7661558 | 0.01 |
ENSRNOT00000080229
|
Fam126a
|
family with sequence similarity 126, member A |
| chr9_-_44024876 | 0.01 |
ENSRNOT00000024366
|
Coa5
|
cytochrome C oxidase assembly factor 5 |
| chr10_+_70411738 | 0.01 |
ENSRNOT00000078046
|
Slfn4
|
schlafen 4 |
| chr1_-_141893674 | 0.01 |
ENSRNOT00000019059
ENSRNOT00000085988 |
Idh2
|
isocitrate dehydrogenase (NADP(+)) 2, mitochondrial |
| chrX_+_34623405 | 0.01 |
ENSRNOT00000041924
|
Nhs
|
NHS actin remodeling regulator |
| chr15_+_30557388 | 0.01 |
ENSRNOT00000072978
|
AABR07017745.4
|
|
| chrX_+_69730242 | 0.01 |
ENSRNOT00000075980
ENSRNOT00000076425 |
Eda
|
ectodysplasin-A |
| chr3_+_147511563 | 0.01 |
ENSRNOT00000006828
|
Slc52a3
|
solute carrier family 52 member 3 |
| chr3_+_56802714 | 0.01 |
ENSRNOT00000077551
|
Erich2
|
glutamate-rich 2 |
| chr3_+_114253637 | 0.01 |
ENSRNOT00000046843
|
Duox1
|
dual oxidase 1 |
| chr5_+_98387291 | 0.01 |
ENSRNOT00000046503
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr4_-_23718047 | 0.01 |
ENSRNOT00000011359
|
RGD1564345
|
similar to RIKEN cDNA 4921511H03 |
| chr1_+_143583675 | 0.01 |
ENSRNOT00000026385
|
Fam103a1
|
family with sequence similarity 103, member A1 |
| chr3_-_123236535 | 0.01 |
ENSRNOT00000028839
|
Slc4a11
|
solute carrier family 4 member 11 |
| chr10_+_37032043 | 0.01 |
ENSRNOT00000037303
ENSRNOT00000091242 |
Phykpl
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr15_+_57290849 | 0.01 |
ENSRNOT00000014909
|
Cpb2
|
carboxypeptidase B2 |
| chr10_-_110232843 | 0.01 |
ENSRNOT00000054934
|
Cd7
|
Cd7 molecule |
| chr7_+_125920357 | 0.01 |
ENSRNOT00000018365
|
Upk3a
|
uroplakin 3A |
| chr3_-_2453933 | 0.01 |
ENSRNOT00000014060
|
Slc34a3
|
solute carrier family 34 member 3 |
| chr15_-_37410848 | 0.01 |
ENSRNOT00000081757
|
Gjb6
|
gap junction protein, beta 6 |
| chr1_+_234252757 | 0.01 |
ENSRNOT00000091814
|
Rorb
|
RAR-related orphan receptor B |
| chr18_+_44716226 | 0.01 |
ENSRNOT00000086431
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chr1_-_175895510 | 0.01 |
ENSRNOT00000064535
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr13_+_82072497 | 0.01 |
ENSRNOT00000063810
ENSRNOT00000085135 |
Kifap3
|
kinesin-associated protein 3 |
| chr19_+_10024947 | 0.01 |
ENSRNOT00000061392
|
Cfap20
|
cilia and flagella associated protein 20 |
| chr20_+_31761405 | 0.01 |
ENSRNOT00000000661
|
Neurog3
|
neurogenin 3 |
| chr1_+_63263722 | 0.01 |
ENSRNOT00000088203
|
LOC100911184
|
vomeronasal type-1 receptor 4-like |
| chr14_+_44889287 | 0.01 |
ENSRNOT00000091312
ENSRNOT00000032273 |
Tmem156
|
transmembrane protein 156 |
| chr1_-_72941869 | 0.01 |
ENSRNOT00000032306
|
Eps8l1
|
EPS8-like 1 |
| chr17_-_84247038 | 0.01 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tal1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1901423 | response to benzene(GO:1901423) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.1 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.0 | 0.1 | GO:0072249 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) |
| 0.0 | 0.0 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.2 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.0 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.1 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.0 | GO:2000814 | positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.0 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.0 | GO:0032218 | riboflavin transport(GO:0032218) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0048237 | Weibel-Palade body(GO:0033093) rough endoplasmic reticulum lumen(GO:0048237) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.0 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |