Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
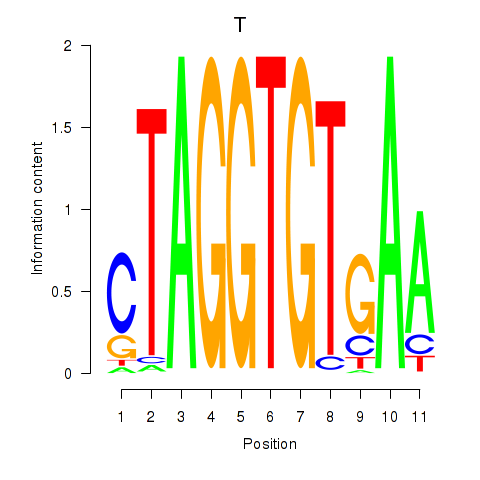
Results for T
Z-value: 0.33
Transcription factors associated with T
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
T
|
ENSRNOG00000012229 | T brachyury transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| T | rn6_v1_chr1_-_52894832_52894832 | 0.20 | 7.4e-01 | Click! |
Activity profile of T motif
Sorted Z-values of T motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_152883210 | 0.22 |
ENSRNOT00000056200
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr4_-_148845267 | 0.16 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr1_-_219422268 | 0.12 |
ENSRNOT00000025092
|
Carns1
|
carnosine synthase 1 |
| chr8_-_46694613 | 0.11 |
ENSRNOT00000084514
|
AC133265.1
|
|
| chr7_-_130120579 | 0.10 |
ENSRNOT00000044376
|
Mapk12
|
mitogen-activated protein kinase 12 |
| chr2_-_23289266 | 0.09 |
ENSRNOT00000061708
|
Bhmt2
|
betaine-homocysteine S-methyltransferase 2 |
| chr13_-_25262469 | 0.09 |
ENSRNOT00000019921
|
Rnf152
|
ring finger protein 152 |
| chr8_+_72405748 | 0.08 |
ENSRNOT00000023952
|
Car12
|
carbonic anhydrase 12 |
| chr9_-_20528879 | 0.07 |
ENSRNOT00000085293
|
AABR07066871.3
|
|
| chr4_-_59014416 | 0.07 |
ENSRNOT00000049811
|
RGD1561636
|
similar to 60S ribosomal protein L38 |
| chr10_+_55712043 | 0.06 |
ENSRNOT00000010141
|
Aloxe3
|
arachidonate lipoxygenase 3 |
| chr15_+_34552410 | 0.06 |
ENSRNOT00000027802
|
Khnyn
|
KH and NYN domain containing |
| chr1_-_214423881 | 0.06 |
ENSRNOT00000025290
|
Pidd1
|
p53-induced death domain protein 1 |
| chr5_-_62186372 | 0.06 |
ENSRNOT00000089789
|
Coro2a
|
coronin 2A |
| chr18_+_38847632 | 0.06 |
ENSRNOT00000014916
|
LOC102554034
|
ELMO domain-containing protein 2-like |
| chr7_-_47586137 | 0.05 |
ENSRNOT00000006086
|
Tmtc2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr9_-_89193821 | 0.05 |
ENSRNOT00000090881
|
Sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr15_+_10120206 | 0.05 |
ENSRNOT00000033048
|
Rarb
|
retinoic acid receptor, beta |
| chr20_-_27578244 | 0.05 |
ENSRNOT00000000708
|
Fam26d
|
family with sequence similarity 26, member D |
| chr18_-_3662654 | 0.05 |
ENSRNOT00000092854
ENSRNOT00000016167 |
Npc1
|
NPC intracellular cholesterol transporter 1 |
| chr14_-_16996962 | 0.05 |
ENSRNOT00000091936
|
AC103535.1
|
|
| chr5_-_173233188 | 0.04 |
ENSRNOT00000055343
|
Tmem88b
|
transmembrane protein 88B |
| chr2_-_9504134 | 0.04 |
ENSRNOT00000076996
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr8_-_58195884 | 0.04 |
ENSRNOT00000010573
|
Acat1
|
acetyl-CoA acetyltransferase 1 |
| chr10_-_61772250 | 0.04 |
ENSRNOT00000092338
ENSRNOT00000085394 ENSRNOT00000046110 |
Srr
|
serine racemase |
| chr1_-_282492912 | 0.04 |
ENSRNOT00000047883
|
AABR07007130.1
|
|
| chr10_-_73865364 | 0.03 |
ENSRNOT00000005226
|
Rps6kb1
|
ribosomal protein S6 kinase B1 |
| chr2_-_206274079 | 0.03 |
ENSRNOT00000056079
|
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr14_+_83510278 | 0.03 |
ENSRNOT00000081161
|
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr3_+_161413298 | 0.03 |
ENSRNOT00000023965
ENSRNOT00000088776 |
Mmp9
|
matrix metallopeptidase 9 |
| chr10_+_35637133 | 0.03 |
ENSRNOT00000043085
|
Tbc1d9b
|
TBC1 domain family member 9B |
| chr10_+_61772449 | 0.03 |
ENSRNOT00000004058
|
Smg6
|
SMG6 nonsense mediated mRNA decay factor |
| chr6_+_72092194 | 0.03 |
ENSRNOT00000005670
|
G2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr4_+_145282797 | 0.03 |
ENSRNOT00000085766
|
Ogg1
|
8-oxoguanine DNA glycosylase |
| chr14_-_87701884 | 0.03 |
ENSRNOT00000079338
|
Mospd1
|
motile sperm domain containing 1 |
| chr5_+_117586103 | 0.03 |
ENSRNOT00000084640
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr11_+_73198522 | 0.03 |
ENSRNOT00000002356
|
Xxylt1
|
xyloside xylosyltransferase 1 |
| chr13_+_56513286 | 0.03 |
ENSRNOT00000015596
|
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chrX_+_33599671 | 0.03 |
ENSRNOT00000006843
|
Txlng
|
taxilin gamma |
| chr6_-_127508452 | 0.03 |
ENSRNOT00000073709
|
LOC100909524
|
protein Z-dependent protease inhibitor-like |
| chr2_-_188596222 | 0.03 |
ENSRNOT00000027920
|
Efna1
|
ephrin A1 |
| chrX_-_75566481 | 0.03 |
ENSRNOT00000003714
|
Zdhhc15
|
zinc finger, DHHC-type containing 15 |
| chr12_+_17253791 | 0.03 |
ENSRNOT00000083814
|
Zfand2a
|
zinc finger AN1-type containing 2A |
| chr3_-_164728338 | 0.03 |
ENSRNOT00000035463
|
Fam65c
|
family with sequence similarity 65, member C |
| chr10_+_15183803 | 0.03 |
ENSRNOT00000039777
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr1_+_116604550 | 0.03 |
ENSRNOT00000083587
|
Ube3a
|
ubiquitin protein ligase E3A |
| chr1_-_89559960 | 0.02 |
ENSRNOT00000092133
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chrX_-_105793306 | 0.02 |
ENSRNOT00000015806
|
Nxf2
|
nuclear RNA export factor 2 |
| chr18_+_28361283 | 0.02 |
ENSRNOT00000026949
|
Matr3
|
matrin 3 |
| chr9_-_19372673 | 0.02 |
ENSRNOT00000073667
ENSRNOT00000079517 |
Clic5
|
chloride intracellular channel 5 |
| chr1_+_238222521 | 0.02 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr10_+_38788992 | 0.02 |
ENSRNOT00000009727
|
Gdf9
|
growth differentiation factor 9 |
| chr2_+_68821004 | 0.02 |
ENSRNOT00000083713
|
Egf
|
epidermal growth factor |
| chr6_+_129835919 | 0.02 |
ENSRNOT00000036035
|
Vrk1
|
vaccinia related kinase 1 |
| chr5_+_148320438 | 0.02 |
ENSRNOT00000018742
|
Pef1
|
penta-EF hand domain containing 1 |
| chr20_-_8574082 | 0.02 |
ENSRNOT00000048845
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr10_+_93811505 | 0.02 |
ENSRNOT00000081937
|
Tanc2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr11_+_88699222 | 0.02 |
ENSRNOT00000084177
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr3_-_133894569 | 0.02 |
ENSRNOT00000084221
|
AABR07053968.1
|
|
| chr1_+_128199322 | 0.02 |
ENSRNOT00000075131
|
Lysmd4
|
LysM domain containing 4 |
| chr10_-_56289882 | 0.02 |
ENSRNOT00000090762
ENSRNOT00000056903 |
Tnfsf13
|
tumor necrosis factor superfamily member 13 |
| chr1_+_197839430 | 0.02 |
ENSRNOT00000025043
|
Rabep2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr11_-_55240241 | 0.02 |
ENSRNOT00000064614
|
Dppa4
|
developmental pluripotency associated 4 |
| chr1_-_141533908 | 0.02 |
ENSRNOT00000020117
|
Mesp1
|
mesoderm posterior bHLH transcription factor 1 |
| chr4_+_122365093 | 0.02 |
ENSRNOT00000024011
|
Klf15
|
Kruppel-like factor 15 |
| chr4_-_170195107 | 0.02 |
ENSRNOT00000073808
|
LOC100912450
|
uncharacterized LOC100912450 |
| chr4_-_170813012 | 0.02 |
ENSRNOT00000073975
|
LOC108348150
|
single-pass membrane and coiled-coil domain-containing protein 3 |
| chrX_-_111179152 | 0.02 |
ENSRNOT00000089115
|
Morc4
|
MORC family CW-type zinc finger 4 |
| chr1_-_213635546 | 0.02 |
ENSRNOT00000018861
|
Sirt3
|
sirtuin 3 |
| chr2_-_1511591 | 0.02 |
ENSRNOT00000087906
ENSRNOT00000014709 |
Cast
|
calpastatin |
| chr10_+_63778755 | 0.01 |
ENSRNOT00000090481
|
Inpp5k
|
inositol polyphosphate-5-phosphatase K |
| chr20_+_47494270 | 0.01 |
ENSRNOT00000036365
|
Sec63
|
SEC63 homolog, protein translocation regulator |
| chr20_+_6356423 | 0.01 |
ENSRNOT00000000628
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr19_-_37907714 | 0.01 |
ENSRNOT00000026361
|
Ctrl
|
chymotrypsin-like |
| chr18_-_69723869 | 0.01 |
ENSRNOT00000000125
|
Elac1
|
elaC ribonuclease Z 1 |
| chr13_+_49092306 | 0.01 |
ENSRNOT00000000039
|
Nuak2
|
NUAK family kinase 2 |
| chr1_+_213451721 | 0.01 |
ENSRNOT00000045652
|
Olr315
|
olfactory receptor 315 |
| chr10_-_51778939 | 0.01 |
ENSRNOT00000078675
ENSRNOT00000057562 |
Myocd
|
myocardin |
| chr3_+_149493655 | 0.01 |
ENSRNOT00000018226
|
Bpifa2
|
BPI fold containing family A, member 2 |
| chr10_+_11206226 | 0.01 |
ENSRNOT00000006979
|
Tfap4
|
transcription factor AP-4 |
| chr10_-_47630799 | 0.01 |
ENSRNOT00000057907
|
Slc47a2
|
solute carrier family 47 member 2 |
| chr3_+_175493698 | 0.01 |
ENSRNOT00000087356
|
Osbpl2
|
oxysterol binding protein-like 2 |
| chr15_+_107486555 | 0.01 |
ENSRNOT00000074277
|
AABR07019512.1
|
|
| chr1_-_167698263 | 0.01 |
ENSRNOT00000093046
|
Trim21
|
tripartite motif-containing 21 |
| chr13_+_89774764 | 0.01 |
ENSRNOT00000005619
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr17_+_10066187 | 0.01 |
ENSRNOT00000091200
|
Uimc1
|
ubiquitin interaction motif containing 1 |
| chr6_-_103605256 | 0.01 |
ENSRNOT00000006005
|
Dcaf5
|
DDB1 and CUL4 associated factor 5 |
| chr19_+_39126589 | 0.01 |
ENSRNOT00000027561
|
Sntb2
|
syntrophin, beta 2 |
| chr1_-_215858034 | 0.01 |
ENSRNOT00000027656
|
Ins2
|
insulin 2 |
| chr4_-_78074906 | 0.01 |
ENSRNOT00000080654
|
Zfp467
|
zinc finger protein 467 |
| chr4_+_142780987 | 0.01 |
ENSRNOT00000056570
|
Grm7
|
glutamate metabotropic receptor 7 |
| chr5_-_62001196 | 0.01 |
ENSRNOT00000012602
|
Trmo
|
tRNA methyltransferase O |
| chr12_+_46869836 | 0.01 |
ENSRNOT00000084421
|
Sirt4
|
sirtuin 4 |
| chr1_+_213636093 | 0.01 |
ENSRNOT00000019642
|
Psmd13
|
proteasome 26S subunit, non-ATPase 13 |
| chr8_-_124399494 | 0.01 |
ENSRNOT00000037883
|
Tgfbr2
|
transforming growth factor, beta receptor 2 |
| chr16_-_18645941 | 0.01 |
ENSRNOT00000079241
|
Dydc2
|
DPY30 domain containing 2 |
| chr4_+_89079014 | 0.01 |
ENSRNOT00000087451
|
Herc3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr14_-_14390699 | 0.01 |
ENSRNOT00000046639
|
Anxa3
|
annexin A3 |
| chr4_-_32087600 | 0.01 |
ENSRNOT00000013841
|
Sem1
|
SEM1, 26S proteasome complex subunit |
| chr4_-_78075398 | 0.01 |
ENSRNOT00000010377
|
Zfp467
|
zinc finger protein 467 |
| chr10_-_109966782 | 0.01 |
ENSRNOT00000078052
ENSRNOT00000075041 |
Rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_+_189870622 | 0.01 |
ENSRNOT00000075003
|
Tmem159
|
transmembrane protein 159 |
| chr11_+_74057361 | 0.01 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chrX_+_54266687 | 0.01 |
ENSRNOT00000058309
|
LOC100363193
|
LRRGT00076-like |
| chr1_+_220243473 | 0.01 |
ENSRNOT00000084650
ENSRNOT00000027091 |
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr7_-_99677268 | 0.01 |
ENSRNOT00000013315
|
RGD1564420
|
similar to Hypothetical protein MGC31278 |
| chr16_+_20962227 | 0.01 |
ENSRNOT00000027615
|
Slc25a42
|
solute carrier family 25, member 42 |
| chr16_+_46731403 | 0.01 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr1_-_73121085 | 0.01 |
ENSRNOT00000073464
|
LOC684545
|
similar to NACHT, leucine rich repeat and PYD containing 2 |
| chr9_-_81565416 | 0.00 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chr10_+_32824 | 0.00 |
ENSRNOT00000077828
|
LOC103689943
|
meiosis arrest female protein 1-like |
| chr1_+_193335645 | 0.00 |
ENSRNOT00000019640
|
Lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr12_+_40466495 | 0.00 |
ENSRNOT00000001816
|
Aldh2
|
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr16_-_81834945 | 0.00 |
ENSRNOT00000037806
|
F7
|
coagulation factor VII |
| chr3_+_151126591 | 0.00 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr3_-_176706896 | 0.00 |
ENSRNOT00000017337
|
Ptk6
|
protein tyrosine kinase 6 |
| chr1_-_133559975 | 0.00 |
ENSRNOT00000046645
|
Mctp2
|
multiple C2 and transmembrane domain containing 2 |
| chr4_+_118243132 | 0.00 |
ENSRNOT00000023654
|
RGD1306746
|
similar to Hypothetical protein MGC25529 |
| chr7_+_78732904 | 0.00 |
ENSRNOT00000088433
|
LOC103690326
|
dendritic cell-specific transmembrane protein |
| chr4_-_9060554 | 0.00 |
ENSRNOT00000046248
|
AABR07059215.1
|
|
| chr9_-_63637677 | 0.00 |
ENSRNOT00000049259
|
Satb2
|
SATB homeobox 2 |
| chrX_+_159703578 | 0.00 |
ENSRNOT00000001162
|
Cd40lg
|
CD40 ligand |
| chr6_-_21112734 | 0.00 |
ENSRNOT00000079819
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chr2_+_43271092 | 0.00 |
ENSRNOT00000017571
|
Mier3
|
mesoderm induction early response 1, family member 3 |
| chr1_-_22404002 | 0.00 |
ENSRNOT00000044098
|
Taar8a
|
trace amine-associated receptor 8a |
| chr5_-_171415354 | 0.00 |
ENSRNOT00000055398
|
Tp73
|
tumor protein p73 |
| chr1_-_240601744 | 0.00 |
ENSRNOT00000024093
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr8_-_75995371 | 0.00 |
ENSRNOT00000014014
|
Foxb1
|
forkhead box B1 |
| chr19_+_37600148 | 0.00 |
ENSRNOT00000023853
|
Ctcf
|
CCCTC-binding factor |
| chr3_+_122928964 | 0.00 |
ENSRNOT00000074724
|
LOC100365450
|
rCG26795-like |
| chr16_-_82999827 | 0.00 |
ENSRNOT00000041223
|
Tex29
|
testis expressed 29 |
Network of associatons between targets according to the STRING database.
First level regulatory network of T
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0046952 | acetyl-CoA catabolic process(GO:0046356) ketone body catabolic process(GO:0046952) metanephric proximal tubule development(GO:0072237) |
| 0.0 | 0.0 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.0 | GO:0071283 | response to high light intensity(GO:0009644) cellular response to iron(III) ion(GO:0071283) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |