Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
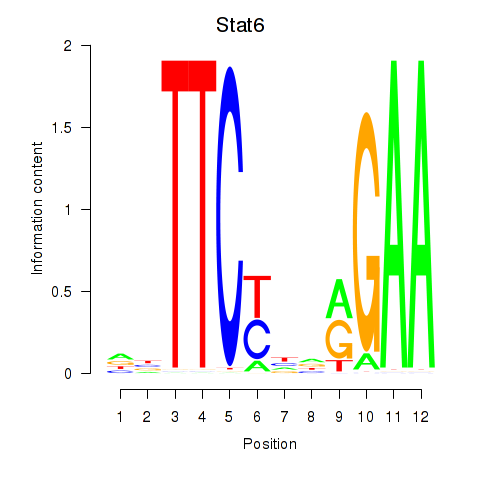
Results for Stat6
Z-value: 0.39
Transcription factors associated with Stat6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat6
|
ENSRNOG00000025023 | signal transducer and activator of transcription 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat6 | rn6_v1_chr7_+_70946228_70946228 | 0.90 | 3.9e-02 | Click! |
Activity profile of Stat6 motif
Sorted Z-values of Stat6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_173087648 | 0.17 |
ENSRNOT00000091079
|
Iqcj
|
IQ motif containing J |
| chr5_-_138697641 | 0.15 |
ENSRNOT00000012062
|
Guca2b
|
guanylate cyclase activator 2B |
| chr4_-_22425381 | 0.13 |
ENSRNOT00000011166
ENSRNOT00000037712 |
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr18_+_30913842 | 0.13 |
ENSRNOT00000026947
|
Pcdhgc3
|
protocadherin gamma subfamily C, 3 |
| chr3_+_110723481 | 0.13 |
ENSRNOT00000013878
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr13_-_109580271 | 0.12 |
ENSRNOT00000088326
|
Flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr5_-_166116516 | 0.12 |
ENSRNOT00000079919
ENSRNOT00000080888 |
Kif1b
|
kinesin family member 1B |
| chr3_-_158328881 | 0.12 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr9_+_111028824 | 0.11 |
ENSRNOT00000041418
ENSRNOT00000056457 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr15_+_83703791 | 0.11 |
ENSRNOT00000090637
|
Klf5
|
Kruppel-like factor 5 |
| chr13_-_88642011 | 0.11 |
ENSRNOT00000067037
|
LOC100361087
|
hypothetical LOC100361087 |
| chr4_-_40036563 | 0.10 |
ENSRNOT00000084531
|
Tmem168
|
transmembrane protein 168 |
| chr4_+_66090209 | 0.10 |
ENSRNOT00000031193
|
Ttc26
|
tetratricopeptide repeat domain 26 |
| chr8_-_49158971 | 0.10 |
ENSRNOT00000020573
|
Kmt2a
|
lysine methyltransferase 2A |
| chr14_+_39661968 | 0.10 |
ENSRNOT00000064779
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr7_+_97559841 | 0.09 |
ENSRNOT00000007326
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr1_+_218076116 | 0.09 |
ENSRNOT00000028374
|
Oraov1
|
oral cancer overexpressed 1 |
| chr10_+_106712127 | 0.08 |
ENSRNOT00000040629
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr2_+_149899836 | 0.08 |
ENSRNOT00000086481
|
RGD1560324
|
similar to hypothetical protein C130079G13 |
| chr2_+_189062443 | 0.08 |
ENSRNOT00000028181
|
Adar
|
adenosine deaminase, RNA-specific |
| chr19_-_22281778 | 0.08 |
ENSRNOT00000049624
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr7_+_141666111 | 0.08 |
ENSRNOT00000083250
|
AABR07058886.1
|
|
| chr19_+_38768467 | 0.08 |
ENSRNOT00000027346
|
Cdh1
|
cadherin 1 |
| chr1_-_88066101 | 0.08 |
ENSRNOT00000079473
ENSRNOT00000027893 |
Ryr1
|
ryanodine receptor 1 |
| chrX_-_19386541 | 0.08 |
ENSRNOT00000072722
|
AABR07037356.1
|
|
| chr13_-_47438360 | 0.08 |
ENSRNOT00000067445
|
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr2_+_189629297 | 0.08 |
ENSRNOT00000049810
|
Dennd4b
|
DENN domain containing 4B |
| chr4_-_88565292 | 0.07 |
ENSRNOT00000008948
|
Lancl2
|
LanC like 2 |
| chr3_-_37480984 | 0.07 |
ENSRNOT00000030373
|
Nmi
|
N-myc (and STAT) interactor |
| chrX_+_68891227 | 0.07 |
ENSRNOT00000009635
|
Efnb1
|
ephrin B1 |
| chrX_-_135419704 | 0.07 |
ENSRNOT00000044435
|
Zfp280c
|
zinc finger protein 280C |
| chr8_+_62317265 | 0.07 |
ENSRNOT00000083418
ENSRNOT00000091204 |
Fam219b
|
family with sequence similarity 219, member B |
| chr14_+_37113210 | 0.06 |
ENSRNOT00000089094
|
Sgcb
|
sarcoglycan, beta |
| chr12_-_35979193 | 0.06 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr17_-_15467320 | 0.06 |
ENSRNOT00000072490
ENSRNOT00000093561 |
Ogn
|
osteoglycin |
| chr16_+_11032531 | 0.06 |
ENSRNOT00000078390
|
Wapl
|
WAPL cohesin release factor |
| chr15_+_4209703 | 0.05 |
ENSRNOT00000082236
|
Ppp3cb
|
protein phosphatase 3 catalytic subunit beta |
| chr14_-_28967980 | 0.05 |
ENSRNOT00000048175
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr14_+_7618022 | 0.05 |
ENSRNOT00000088508
ENSRNOT00000002819 |
Slc10a6
|
solute carrier family 10 member 6 |
| chr15_+_45712821 | 0.05 |
ENSRNOT00000083381
ENSRNOT00000045833 |
Fam124a
|
family with sequence similarity 124 member A |
| chr13_+_36156182 | 0.05 |
ENSRNOT00000071437
|
LOC288978
|
hypothetical LOC288978 |
| chr4_-_133951264 | 0.05 |
ENSRNOT00000090506
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr1_-_247985497 | 0.05 |
ENSRNOT00000078998
|
Ranbp6
|
RAN binding protein 6 |
| chr5_-_135994848 | 0.05 |
ENSRNOT00000067675
|
Btbd19
|
BTB domain containing 19 |
| chr18_+_25637588 | 0.05 |
ENSRNOT00000075283
ENSRNOT00000031530 |
Wdr36
|
WD repeat domain 36 |
| chr14_-_80355420 | 0.05 |
ENSRNOT00000049798
|
Acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr8_-_129919120 | 0.05 |
ENSRNOT00000074423
|
Ulk4
|
unc-51 like kinase 4 |
| chr15_-_33725188 | 0.05 |
ENSRNOT00000083941
|
Zfhx2
|
zinc finger homeobox 2 |
| chr14_+_60657686 | 0.05 |
ENSRNOT00000070892
|
Sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr3_-_133122691 | 0.04 |
ENSRNOT00000083637
|
Tasp1
|
taspase 1 |
| chr13_+_90184569 | 0.04 |
ENSRNOT00000082393
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr1_+_171793782 | 0.04 |
ENSRNOT00000081351
|
Olfml1
|
olfactomedin-like 1 |
| chr5_-_101405656 | 0.04 |
ENSRNOT00000058611
|
Ttc39b
|
tetratricopeptide repeat domain 39B |
| chr5_+_116421894 | 0.04 |
ENSRNOT00000080577
ENSRNOT00000086628 ENSRNOT00000004017 |
Nfia
|
nuclear factor I/A |
| chr1_+_88582641 | 0.04 |
ENSRNOT00000028187
|
Zfp382
|
zinc finger protein 382 |
| chr3_-_21697945 | 0.04 |
ENSRNOT00000012433
|
Zbtb26
|
zinc finger and BTB domain containing 26 |
| chr1_+_221710670 | 0.04 |
ENSRNOT00000064798
|
Map4k2
|
mitogen activated protein kinase kinase kinase kinase 2 |
| chr1_+_198199622 | 0.04 |
ENSRNOT00000026688
|
Gdpd3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr1_-_37932084 | 0.04 |
ENSRNOT00000022527
|
LOC102547287
|
zinc finger protein 728-like |
| chr2_+_257633425 | 0.04 |
ENSRNOT00000071770
|
Zzz3
|
zinc finger, ZZ-type containing 3 |
| chr6_+_135549137 | 0.04 |
ENSRNOT00000073989
|
Rcor1
|
REST corepressor 1 |
| chr18_+_30820321 | 0.04 |
ENSRNOT00000060472
|
Pcdhga3
|
protocadherin gamma subfamily A, 3 |
| chr20_+_3300247 | 0.04 |
ENSRNOT00000035838
|
Prr3
|
proline rich 3 |
| chr18_+_16146447 | 0.04 |
ENSRNOT00000022117
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr2_+_185590986 | 0.04 |
ENSRNOT00000088807
ENSRNOT00000088188 |
Lrba
|
LPS responsive beige-like anchor protein |
| chr3_-_150960030 | 0.04 |
ENSRNOT00000024714
|
Ncoa6
|
nuclear receptor coactivator 6 |
| chr18_+_30840868 | 0.04 |
ENSRNOT00000027026
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr4_+_78354335 | 0.03 |
ENSRNOT00000059167
|
Gimap7
|
GTPase, IMAP family member 7 |
| chr7_-_70467915 | 0.03 |
ENSRNOT00000088995
|
Slc26a10
|
solute carrier family 26, member 10 |
| chrX_+_20423401 | 0.03 |
ENSRNOT00000093162
|
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr1_-_6970040 | 0.03 |
ENSRNOT00000016273
|
Utrn
|
utrophin |
| chr15_+_34282936 | 0.03 |
ENSRNOT00000026364
|
Irf9
|
interferon regulatory factor 9 |
| chr8_+_45797315 | 0.03 |
ENSRNOT00000059997
|
AABR07070046.1
|
|
| chr4_+_183896303 | 0.03 |
ENSRNOT00000055437
|
RGD1309621
|
similar to hypothetical protein FLJ10652 |
| chr4_-_148437961 | 0.03 |
ENSRNOT00000082907
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr15_-_12513931 | 0.03 |
ENSRNOT00000010103
|
Atxn7
|
ataxin 7 |
| chrX_-_15504165 | 0.03 |
ENSRNOT00000006233
|
Otud5
|
OTU deubiquitinase 5 |
| chr3_+_21698032 | 0.03 |
ENSRNOT00000013378
|
Rabgap1
|
RAB GTPase activating protein 1 |
| chr8_-_36374673 | 0.03 |
ENSRNOT00000013763
|
Dcps
|
decapping enzyme, scavenger |
| chr10_-_89130339 | 0.02 |
ENSRNOT00000027640
|
Ezh1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr4_-_58881045 | 0.02 |
ENSRNOT00000087241
|
Podxl
|
podocalyxin-like |
| chr13_+_48790767 | 0.02 |
ENSRNOT00000087504
|
Elk4
|
ELK4, ETS transcription factor |
| chr3_+_66593934 | 0.02 |
ENSRNOT00000008046
|
Ssfa2
|
sperm specific antigen 2 |
| chr15_+_41937880 | 0.02 |
ENSRNOT00000032514
|
Kcnrg
|
potassium channel regulator |
| chrX_+_9815652 | 0.02 |
ENSRNOT00000091382
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr2_+_8732932 | 0.02 |
ENSRNOT00000072849
|
Arrdc3
|
arrestin domain containing 3 |
| chr1_+_99486253 | 0.02 |
ENSRNOT00000074820
|
LOC102546648
|
uncharacterized LOC102546648 |
| chr1_+_162549420 | 0.02 |
ENSRNOT00000037050
|
Rsf1
|
remodeling and spacing factor 1 |
| chr11_+_7265828 | 0.02 |
ENSRNOT00000084765
|
Gbe1
|
1,4-alpha-glucan branching enzyme 1 |
| chr9_-_14668297 | 0.02 |
ENSRNOT00000042404
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr11_+_60054408 | 0.02 |
ENSRNOT00000091756
|
Abhd10
|
abhydrolase domain containing 10 |
| chr7_-_60341264 | 0.02 |
ENSRNOT00000007747
|
Lyz2
|
lysozyme 2 |
| chrX_+_9436707 | 0.02 |
ENSRNOT00000004187
|
Cask
|
calcium/calmodulin dependent serine protein kinase |
| chr8_+_70789256 | 0.02 |
ENSRNOT00000048302
|
Clpx
|
caseinolytic mitochondrial matrix peptidase chaperone subunit |
| chr9_+_10941613 | 0.02 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr1_-_165123455 | 0.02 |
ENSRNOT00000024875
|
Pold3
|
DNA polymerase delta 3, accessory subunit |
| chr14_+_75880410 | 0.01 |
ENSRNOT00000014078
|
Hs3st1
|
heparan sulfate-glucosamine 3-sulfotransferase 1 |
| chr1_+_143036218 | 0.01 |
ENSRNOT00000025797
|
Pde8a
|
phosphodiesterase 8A |
| chr4_-_10329241 | 0.01 |
ENSRNOT00000017232
|
Fgl2
|
fibrinogen-like 2 |
| chr10_-_56368360 | 0.01 |
ENSRNOT00000019674
|
Slc35g3
|
solute carrier family 35, member G3 |
| chr3_-_63568464 | 0.01 |
ENSRNOT00000068494
|
AABR07052585.2
|
|
| chr1_+_260153645 | 0.01 |
ENSRNOT00000054717
|
Zfp518a
|
zinc finger protein 518A |
| chr20_+_1736377 | 0.01 |
ENSRNOT00000047035
|
Olr1734
|
olfactory receptor 1734 |
| chr16_+_23447366 | 0.01 |
ENSRNOT00000068629
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr10_+_63677396 | 0.01 |
ENSRNOT00000005100
|
Slc43a2
|
solute carrier family 43 member 2 |
| chr15_+_61826937 | 0.01 |
ENSRNOT00000084005
ENSRNOT00000079417 |
Elf1
|
E74-like factor 1 |
| chr11_-_54344615 | 0.01 |
ENSRNOT00000090307
|
Myh15
|
myosin, heavy chain 15 |
| chr1_-_124803363 | 0.01 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr10_+_75087892 | 0.01 |
ENSRNOT00000065910
|
Mpo
|
myeloperoxidase |
| chr3_+_56862691 | 0.01 |
ENSRNOT00000087712
|
Gad1
|
glutamate decarboxylase 1 |
| chr8_-_53816447 | 0.01 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr8_-_114449956 | 0.01 |
ENSRNOT00000056414
|
Col6a6
|
collagen type VI alpha 6 chain |
| chr3_-_147479472 | 0.01 |
ENSRNOT00000074295
|
Fam110a
|
family with sequence similarity 110, member A |
| chr1_-_112947399 | 0.01 |
ENSRNOT00000093306
ENSRNOT00000093259 |
Gabra5
|
gamma-aminobutyric acid type A receptor alpha 5 subunit |
| chr5_-_135561914 | 0.01 |
ENSRNOT00000023178
|
Mmachc
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr8_+_107499262 | 0.01 |
ENSRNOT00000081029
|
Cep70
|
centrosomal protein 70 |
| chr8_+_63036976 | 0.01 |
ENSRNOT00000011383
|
Stoml1
|
stomatin like 1 |
| chr1_-_217844957 | 0.01 |
ENSRNOT00000090653
|
Ano1
|
anoctamin 1 |
| chr3_+_172550258 | 0.01 |
ENSRNOT00000075148
|
Tubb1
|
tubulin, beta 1 class VI |
| chr3_-_5575136 | 0.01 |
ENSRNOT00000008034
|
Slc2a6
|
solute carrier family 2 member 6 |
| chr13_+_100980574 | 0.00 |
ENSRNOT00000067005
ENSRNOT00000004649 |
Capn8
|
calpain 8 |
| chr1_-_103323476 | 0.00 |
ENSRNOT00000019051
|
Mrgprx3
|
MAS related GPR family member X3 |
| chr20_+_5057701 | 0.00 |
ENSRNOT00000001119
|
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C |
| chr18_-_6782757 | 0.00 |
ENSRNOT00000068150
|
Aqp4
|
aquaporin 4 |
| chr1_-_143392532 | 0.00 |
ENSRNOT00000026089
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr10_-_6870011 | 0.00 |
ENSRNOT00000003439
|
RGD1309748
|
similar to CG4768-PA |
| chr3_-_138708332 | 0.00 |
ENSRNOT00000010678
|
Rbbp9
|
RB binding protein 9, serine hydrolase |
| chr13_+_48790951 | 0.00 |
ENSRNOT00000090233
|
Elk4
|
ELK4, ETS transcription factor |
| chr7_-_11513581 | 0.00 |
ENSRNOT00000027045
|
Thop1
|
thimet oligopeptidase 1 |
| chr20_+_2058197 | 0.00 |
ENSRNOT00000077774
ENSRNOT00000040634 |
RT1-M6-1
|
RT1 class I, locus M6, gene 1 |
| chr10_-_37130921 | 0.00 |
ENSRNOT00000006006
|
Trappc2b
|
trafficking protein particle complex 2B |
| chr16_+_7103998 | 0.00 |
ENSRNOT00000016581
|
Pbrm1
|
polybromo 1 |
| chr4_+_62380914 | 0.00 |
ENSRNOT00000029845
|
Tmem140
|
transmembrane protein 140 |
| chrX_-_118513061 | 0.00 |
ENSRNOT00000043338
|
Il13ra2
|
interleukin 13 receptor subunit alpha 2 |
| chr1_+_64114721 | 0.00 |
ENSRNOT00000080466
|
Tmc4
|
transmembrane channel-like 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.1 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.0 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016842 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |