Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
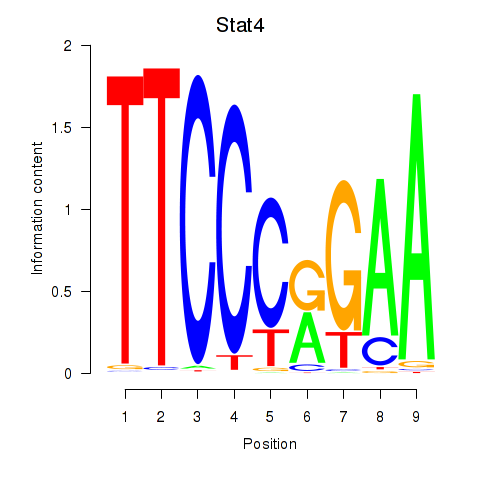
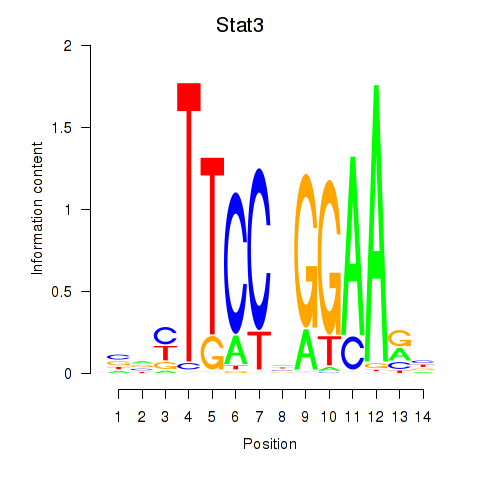
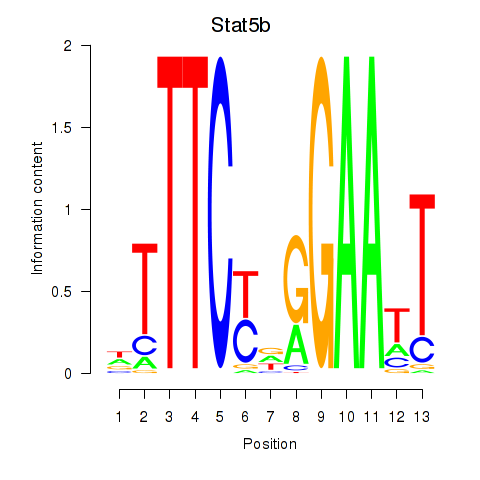
Results for Stat4_Stat3_Stat5b
Z-value: 0.67
Transcription factors associated with Stat4_Stat3_Stat5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat3
|
ENSRNOG00000019742 | signal transducer and activator of transcription 3 |
|
Stat5b
|
ENSRNOG00000019075 | signal transducer and activator of transcription 5B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat3 | rn6_v1_chr10_-_88842233_88842233 | -0.92 | 2.8e-02 | Click! |
| Stat5b | rn6_v1_chr10_-_88712309_88712309 | -0.56 | 3.2e-01 | Click! |
Activity profile of Stat4_Stat3_Stat5b motif
Sorted Z-values of Stat4_Stat3_Stat5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_140090922 | 0.39 |
ENSRNOT00000014456
|
Lalba
|
lactalbumin, alpha |
| chr4_-_23135354 | 0.33 |
ENSRNOT00000011432
|
Steap4
|
STEAP4 metalloreductase |
| chr14_+_63405408 | 0.25 |
ENSRNOT00000086658
|
AABR07015559.1
|
|
| chr2_-_167607919 | 0.24 |
ENSRNOT00000089083
|
AABR07011733.1
|
|
| chr20_-_3793985 | 0.23 |
ENSRNOT00000049540
ENSRNOT00000086293 |
RT1-CE16
|
RT1 class I, locus CE16 |
| chr1_-_78180216 | 0.20 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr7_-_145174771 | 0.16 |
ENSRNOT00000055270
|
Glycam1
|
glycosylation dependent cell adhesion molecule 1 |
| chr15_+_43582020 | 0.15 |
ENSRNOT00000013001
|
Pnma2
|
paraneoplastic Ma antigen 2 |
| chrX_-_143453612 | 0.15 |
ENSRNOT00000051319
|
Atp11c
|
ATPase phospholipid transporting 11C |
| chr5_+_101526551 | 0.14 |
ENSRNOT00000015254
|
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr8_-_36764422 | 0.14 |
ENSRNOT00000017330
|
Hyls1
|
HYLS1, centriolar and ciliogenesis associated |
| chr1_+_42170583 | 0.14 |
ENSRNOT00000093104
|
Vip
|
vasoactive intestinal peptide |
| chr11_+_68105369 | 0.14 |
ENSRNOT00000046888
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr1_+_42170281 | 0.13 |
ENSRNOT00000092879
|
Vip
|
vasoactive intestinal peptide |
| chr9_+_81535483 | 0.13 |
ENSRNOT00000077285
|
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr6_-_42616548 | 0.12 |
ENSRNOT00000081433
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr8_-_77979785 | 0.12 |
ENSRNOT00000084038
|
AC132740.1
|
|
| chr1_-_81412251 | 0.12 |
ENSRNOT00000026946
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr2_+_114386019 | 0.11 |
ENSRNOT00000082148
|
AABR07009834.1
|
|
| chr17_-_15467320 | 0.11 |
ENSRNOT00000072490
ENSRNOT00000093561 |
Ogn
|
osteoglycin |
| chrX_-_25590048 | 0.11 |
ENSRNOT00000004873
|
Mid1
|
midline 1 |
| chr2_-_140618405 | 0.11 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr1_-_266830976 | 0.11 |
ENSRNOT00000027450
|
Pcgf6
|
polycomb group ring finger 6 |
| chr11_+_31806618 | 0.11 |
ENSRNOT00000043410
ENSRNOT00000002769 |
Son
|
Son DNA binding protein |
| chr1_+_154131926 | 0.11 |
ENSRNOT00000035257
|
LOC102549852
|
ferritin light chain 1-like |
| chr15_-_12889102 | 0.10 |
ENSRNOT00000061214
ENSRNOT00000084687 |
RGD1306063
|
similar to HT021 |
| chr14_+_84465515 | 0.10 |
ENSRNOT00000035591
|
Osm
|
oncostatin M |
| chr10_-_95262024 | 0.10 |
ENSRNOT00000066487
|
Bptf
|
bromodomain PHD finger transcription factor |
| chr7_+_54980120 | 0.10 |
ENSRNOT00000005690
ENSRNOT00000005773 |
Kcnc2
|
potassium voltage-gated channel subfamily C member 2 |
| chr2_+_182006242 | 0.10 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr19_-_37281933 | 0.10 |
ENSRNOT00000059663
|
Lrrc29
|
leucine rich repeat containing 29 |
| chr1_-_167093560 | 0.10 |
ENSRNOT00000027301
|
Il18bp
|
interleukin 18 binding protein |
| chr11_+_83868655 | 0.09 |
ENSRNOT00000072402
|
NEWGENE_621438
|
thrombopoietin |
| chr10_-_70744315 | 0.09 |
ENSRNOT00000014865
|
Ccl5
|
C-C motif chemokine ligand 5 |
| chr16_+_33373615 | 0.09 |
ENSRNOT00000079157
|
AABR07025316.1
|
|
| chr6_+_108285822 | 0.09 |
ENSRNOT00000015889
|
Vsx2
|
visual system homeobox 2 |
| chr16_+_33373423 | 0.09 |
ENSRNOT00000091672
|
AABR07025316.1
|
|
| chr7_+_83348694 | 0.09 |
ENSRNOT00000006402
|
Eny2
|
ENY2, transcription and export complex 2 subunit |
| chr16_+_20297414 | 0.09 |
ENSRNOT00000025576
|
Slc5a5
|
solute carrier family 5 member 5 |
| chr4_+_118243132 | 0.09 |
ENSRNOT00000023654
|
RGD1306746
|
similar to Hypothetical protein MGC25529 |
| chr10_-_16752205 | 0.09 |
ENSRNOT00000079462
|
Crebrf
|
CREB3 regulatory factor |
| chr2_+_52152536 | 0.09 |
ENSRNOT00000072691
|
AABR07008293.2
|
|
| chr17_+_47721977 | 0.08 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr15_+_64939527 | 0.08 |
ENSRNOT00000045865
|
AABR07018556.1
|
|
| chr3_+_116072294 | 0.08 |
ENSRNOT00000071786
|
AABR07053613.1
|
|
| chr1_-_81152384 | 0.08 |
ENSRNOT00000026263
ENSRNOT00000077305 |
Zfp94
|
zinc finger protein 94 |
| chr1_-_193700673 | 0.08 |
ENSRNOT00000071314
|
Paip2l1
|
polyadenylate-binding protein-interacting protein 2-like 1 |
| chr2_-_236823145 | 0.08 |
ENSRNOT00000073001
|
AABR07013425.1
|
|
| chr6_-_87427153 | 0.08 |
ENSRNOT00000071999
|
AABR07064622.1
|
|
| chr7_-_83348487 | 0.08 |
ENSRNOT00000006685
|
Nudcd1
|
NudC domain containing 1 |
| chr10_+_34277993 | 0.08 |
ENSRNOT00000055872
ENSRNOT00000003343 |
Ifi47
|
interferon gamma inducible protein 47 |
| chr15_+_41937880 | 0.08 |
ENSRNOT00000032514
|
Kcnrg
|
potassium channel regulator |
| chr9_+_15582564 | 0.08 |
ENSRNOT00000020640
|
LOC100912849
|
uncharacterized LOC100912849 |
| chr15_-_42271443 | 0.08 |
ENSRNOT00000085508
|
AABR07018132.1
|
|
| chr3_+_98297554 | 0.08 |
ENSRNOT00000006524
|
Kcna4
|
potassium voltage-gated channel subfamily A member 4 |
| chr3_+_159887036 | 0.07 |
ENSRNOT00000031868
|
R3hdml
|
R3H domain containing-like |
| chrX_+_73392523 | 0.07 |
ENSRNOT00000073827
|
Zfp449
|
zinc finger protein 449 |
| chr5_+_154668179 | 0.07 |
ENSRNOT00000015971
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chrX_+_153677811 | 0.07 |
ENSRNOT00000072808
|
AABR07042361.1
|
|
| chr19_+_37282018 | 0.07 |
ENSRNOT00000021723
|
Tmem208
|
transmembrane protein 208 |
| chr1_+_157403595 | 0.07 |
ENSRNOT00000012781
|
Ccdc90b
|
coiled-coil domain containing 90B |
| chr2_-_113616766 | 0.07 |
ENSRNOT00000016858
ENSRNOT00000074723 |
Tmem212
|
transmembrane protein 212 |
| chr7_-_102298522 | 0.07 |
ENSRNOT00000006273
|
A1bg
|
alpha-1-B glycoprotein |
| chr2_+_140471690 | 0.07 |
ENSRNOT00000017379
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr9_-_29585448 | 0.07 |
ENSRNOT00000075106
|
AABR07067082.1
|
|
| chrX_+_9436707 | 0.07 |
ENSRNOT00000004187
|
Cask
|
calcium/calmodulin dependent serine protein kinase |
| chr15_+_10462749 | 0.07 |
ENSRNOT00000008035
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr13_+_51022681 | 0.07 |
ENSRNOT00000078599
|
Chi3l1
|
chitinase 3 like 1 |
| chr3_+_114918648 | 0.07 |
ENSRNOT00000089204
|
Sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr9_-_86129073 | 0.07 |
ENSRNOT00000093730
|
Cul3
|
cullin 3 |
| chr7_+_96195209 | 0.07 |
ENSRNOT00000073584
|
AABR07057961.1
|
|
| chr1_+_144239020 | 0.07 |
ENSRNOT00000032106
|
Adamtsl3
|
ADAMTS-like 3 |
| chr18_-_15284077 | 0.07 |
ENSRNOT00000046326
|
LOC498826
|
LRRGT00165 |
| chr7_+_2435553 | 0.07 |
ENSRNOT00000065593
|
Prim1
|
primase (DNA) subunit 1 |
| chr4_-_61573003 | 0.07 |
ENSRNOT00000012179
|
Slc35b4
|
solute carrier family 35 member B4 |
| chr2_+_86996497 | 0.07 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr2_-_27364906 | 0.06 |
ENSRNOT00000078639
|
Polk
|
DNA polymerase kappa |
| chr2_-_179704629 | 0.06 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr6_-_86161333 | 0.06 |
ENSRNOT00000072471
|
AABR07064590.1
|
|
| chr20_-_3397039 | 0.06 |
ENSRNOT00000001084
ENSRNOT00000085259 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr1_-_176079125 | 0.06 |
ENSRNOT00000047044
|
RGD1566189
|
similar to ferritin light chain |
| chr14_-_86387606 | 0.06 |
ENSRNOT00000089384
ENSRNOT00000006642 |
Ddx56
|
DEAD-box helicase 56 |
| chr18_+_36829062 | 0.06 |
ENSRNOT00000025467
ENSRNOT00000086979 |
Tcerg1
|
transcription elongation regulator 1 |
| chr2_+_89254154 | 0.06 |
ENSRNOT00000073391
|
AABR07009221.1
|
|
| chr11_-_31238026 | 0.06 |
ENSRNOT00000032366
|
RGD1562726
|
similar to Putative protein C21orf62 homolog |
| chr2_+_4195917 | 0.06 |
ENSRNOT00000093303
|
RGD1560883
|
similar to KIAA0825 protein |
| chr14_-_43855745 | 0.06 |
ENSRNOT00000083155
|
AC112624.1
|
|
| chr4_+_123118468 | 0.06 |
ENSRNOT00000010895
|
Tmem43
|
transmembrane protein 43 |
| chr1_-_239265997 | 0.06 |
ENSRNOT00000037500
|
RGD1359158
|
similar to RIKEN cDNA 1110059E24 |
| chrX_+_70461718 | 0.06 |
ENSRNOT00000078233
ENSRNOT00000003789 |
Kif4a
|
kinesin family member 4A |
| chr8_-_53362006 | 0.06 |
ENSRNOT00000077145
|
LOC100360390
|
claudin 25-like |
| chr18_+_36596585 | 0.06 |
ENSRNOT00000036613
|
Rbm27
|
RNA binding motif protein 27 |
| chr17_-_67945037 | 0.06 |
ENSRNOT00000023033
|
Klf6
|
Kruppel-like factor 6 |
| chr2_+_86996798 | 0.06 |
ENSRNOT00000070821
|
Zfp455
|
zinc finger protein 455 |
| chr13_-_82005741 | 0.06 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr4_-_29092753 | 0.06 |
ENSRNOT00000014810
|
Bet1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr8_-_23148396 | 0.06 |
ENSRNOT00000075237
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chrX_+_82710329 | 0.05 |
ENSRNOT00000074962
|
AABR07039694.1
|
|
| chr1_-_37741709 | 0.05 |
ENSRNOT00000079186
|
Fastkd3
|
FAST kinase domains 3 |
| chr10_-_56506446 | 0.05 |
ENSRNOT00000021357
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr14_-_100184192 | 0.05 |
ENSRNOT00000007044
|
Plek
|
pleckstrin |
| chr1_-_69963592 | 0.05 |
ENSRNOT00000021245
|
Aurkc
|
aurora kinase C |
| chr14_+_23507628 | 0.05 |
ENSRNOT00000037509
|
Uba6
|
ubiquitin-like modifier activating enzyme 6 |
| chr7_+_1206648 | 0.05 |
ENSRNOT00000073689
|
Pros1
|
protein S (alpha) |
| chr13_-_90405358 | 0.05 |
ENSRNOT00000091710
|
Vangl2
|
VANGL planar cell polarity protein 2 |
| chr20_+_5118026 | 0.05 |
ENSRNOT00000060819
|
G4
|
G4 protein |
| chr2_-_189880743 | 0.05 |
ENSRNOT00000018830
|
Snapin
|
SNAP-associated protein |
| chr1_-_225830300 | 0.05 |
ENSRNOT00000072584
ENSRNOT00000027502 |
Scgb2a2
|
secretoglobin, family 2A, member 2 |
| chr9_+_80002045 | 0.05 |
ENSRNOT00000030761
|
Rpl37a
|
ribosomal protein L37a |
| chr2_+_165601007 | 0.05 |
ENSRNOT00000013931
|
Smc4
|
structural maintenance of chromosomes 4 |
| chr7_-_70355619 | 0.05 |
ENSRNOT00000031272
|
Tspan31
|
tetraspanin 31 |
| chr2_+_166475070 | 0.05 |
ENSRNOT00000071111
|
AABR07011700.1
|
|
| chr10_+_65439102 | 0.05 |
ENSRNOT00000016840
|
Tlcd1
|
TLC domain containing 1 |
| chr3_+_173953869 | 0.05 |
ENSRNOT00000091212
|
Fam217b
|
family with sequence similarity 217, member B |
| chr5_+_78384444 | 0.05 |
ENSRNOT00000034978
|
LOC500475
|
similar to hypothetical protein 4933430I17 |
| chr10_-_13004441 | 0.05 |
ENSRNOT00000004848
|
Cldn9
|
claudin 9 |
| chr8_-_117820413 | 0.05 |
ENSRNOT00000075819
|
Tma7
|
translation machinery associated 7 homolog |
| chr7_-_70328148 | 0.05 |
ENSRNOT00000074645
|
Mettl21b
|
methyltransferase like 21B |
| chr12_-_41448668 | 0.05 |
ENSRNOT00000001856
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr1_+_228684136 | 0.05 |
ENSRNOT00000028608
|
Olr337
|
olfactory receptor 337 |
| chr1_-_52894832 | 0.05 |
ENSRNOT00000016471
|
T
|
T brachyury transcription factor |
| chr10_-_90569575 | 0.05 |
ENSRNOT00000074987
|
AABR07030494.1
|
|
| chr9_-_95108739 | 0.05 |
ENSRNOT00000070819
|
Usp40
|
ubiquitin specific peptidase 40 |
| chrX_+_13441558 | 0.05 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chr2_+_120266933 | 0.05 |
ENSRNOT00000015192
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr20_-_48310175 | 0.05 |
ENSRNOT00000000351
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr2_-_165600748 | 0.05 |
ENSRNOT00000013216
|
Ift80
|
intraflagellar transport 80 |
| chrX_+_31984612 | 0.05 |
ENSRNOT00000005181
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr3_-_46726946 | 0.05 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr18_+_30017918 | 0.05 |
ENSRNOT00000079794
|
Pcdha4
|
protocadherin alpha 4 |
| chr2_-_13696426 | 0.05 |
ENSRNOT00000046251
|
Rasa1
|
RAS p21 protein activator 1 |
| chr6_-_55001464 | 0.05 |
ENSRNOT00000006618
|
Ahr
|
aryl hydrocarbon receptor |
| chr20_+_4852496 | 0.05 |
ENSRNOT00000088936
|
Lta
|
lymphotoxin alpha |
| chr2_-_232117134 | 0.05 |
ENSRNOT00000030798
|
Alpk1
|
alpha-kinase 1 |
| chr2_+_189609800 | 0.05 |
ENSRNOT00000089016
|
Slc39a1
|
solute carrier family 39 member 1 |
| chr3_-_12415073 | 0.05 |
ENSRNOT00000022755
|
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr1_-_219519398 | 0.05 |
ENSRNOT00000025546
|
Ssh3
|
slingshot protein phosphatase 3 |
| chr20_+_3979035 | 0.05 |
ENSRNOT00000000529
|
Tap1
|
transporter 1, ATP binding cassette subfamily B member |
| chr10_+_3227160 | 0.05 |
ENSRNOT00000088075
|
Ntan1
|
N-terminal asparagine amidase |
| chr17_-_76287186 | 0.05 |
ENSRNOT00000092452
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr1_-_104157855 | 0.05 |
ENSRNOT00000019311
|
Csrp3
|
cysteine and glycine rich protein 3 |
| chr5_+_2042991 | 0.05 |
ENSRNOT00000050236
|
Eloc
|
elongin C |
| chr1_-_267463694 | 0.04 |
ENSRNOT00000084851
ENSRNOT00000016645 |
Col17a1
|
collagen type XVII alpha 1 chain |
| chr10_+_70370300 | 0.04 |
ENSRNOT00000055957
ENSRNOT00000076536 |
Slfn2
|
schlafen 2 |
| chr14_+_8080275 | 0.04 |
ENSRNOT00000065965
ENSRNOT00000092542 |
Mapk10
|
mitogen activated protein kinase 10 |
| chr3_+_100768637 | 0.04 |
ENSRNOT00000083542
|
Bdnf
|
brain-derived neurotrophic factor |
| chr1_+_29432152 | 0.04 |
ENSRNOT00000019436
ENSRNOT00000083276 |
Trmt11
|
tRNA methyltransferase 11 homolog |
| chr8_+_64531594 | 0.04 |
ENSRNOT00000036798
|
Gramd2
|
GRAM domain containing 2 |
| chr3_+_147073160 | 0.04 |
ENSRNOT00000012690
|
Sdcbp2
|
syndecan binding protein 2 |
| chr5_+_154800226 | 0.04 |
ENSRNOT00000016046
|
Htr1d
|
5-hydroxytryptamine receptor 1D |
| chr3_+_95959703 | 0.04 |
ENSRNOT00000006406
|
Immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr2_+_86971147 | 0.04 |
ENSRNOT00000032909
|
Zfp458
|
zinc finger protein 458 |
| chr6_-_26875376 | 0.04 |
ENSRNOT00000011838
|
Tmem214
|
transmembrane protein 214 |
| chr7_-_3246071 | 0.04 |
ENSRNOT00000044292
|
Ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr13_-_26811904 | 0.04 |
ENSRNOT00000090097
|
Kdsr
|
3-ketodihydrosphingosine reductase |
| chr6_-_88917070 | 0.04 |
ENSRNOT00000000767
|
Mdga2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chrX_+_14994016 | 0.04 |
ENSRNOT00000006365
|
Ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr1_+_225091612 | 0.04 |
ENSRNOT00000036115
|
Ints5
|
integrator complex subunit 5 |
| chr19_-_41567013 | 0.04 |
ENSRNOT00000022757
|
Zfp612
|
zinc finger protein 612 |
| chr15_-_41338284 | 0.04 |
ENSRNOT00000077225
|
Tnfrsf19
|
TNF receptor superfamily member 19 |
| chr1_+_98228573 | 0.04 |
ENSRNOT00000019278
|
LOC102555672
|
zinc finger protein 850-like |
| chr4_-_15505362 | 0.04 |
ENSRNOT00000009763
|
Hgf
|
hepatocyte growth factor |
| chr16_+_7082450 | 0.04 |
ENSRNOT00000013291
|
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr5_-_64789318 | 0.04 |
ENSRNOT00000078957
|
Mrpl50
|
mitochondrial ribosomal protein L50 |
| chr4_-_51586881 | 0.04 |
ENSRNOT00000064983
|
Iqub
|
IQ motif and ubiquitin domain containing |
| chr4_-_89695928 | 0.04 |
ENSRNOT00000039316
|
Gprin3
|
GPRIN family member 3 |
| chr14_-_23604834 | 0.04 |
ENSRNOT00000002760
|
Stap1
|
signal transducing adaptor family member 1 |
| chr1_-_170431073 | 0.04 |
ENSRNOT00000024710
|
Hpx
|
hemopexin |
| chr1_+_85003280 | 0.04 |
ENSRNOT00000057122
|
Fcgbp
|
Fc fragment of IgG binding protein |
| chr8_+_48657795 | 0.04 |
ENSRNOT00000013134
|
Dpagt1
|
dolichyl-phosphate N-acetylglucosaminephosphotransferase 1 |
| chr6_-_108329464 | 0.04 |
ENSRNOT00000016040
|
Abcd4
|
ATP binding cassette subfamily D member 4 |
| chr15_+_4077951 | 0.04 |
ENSRNOT00000085266
|
Myoz1
|
myozenin 1 |
| chr20_+_2194709 | 0.04 |
ENSRNOT00000001017
|
Trim15
|
tripartite motif containing 15 |
| chr7_-_50638798 | 0.04 |
ENSRNOT00000048880
|
Syt1
|
synaptotagmin 1 |
| chr15_+_34452116 | 0.04 |
ENSRNOT00000027647
|
Ltb4r
|
leukotriene B4 receptor |
| chr10_-_70220558 | 0.04 |
ENSRNOT00000041389
ENSRNOT00000076398 ENSRNOT00000076596 |
Rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr18_+_6207280 | 0.04 |
ENSRNOT00000037955
|
Taf4b
|
TATA-box binding protein associated factor 4b |
| chr10_-_70337532 | 0.04 |
ENSRNOT00000055963
|
Slfn13
|
schlafen family member 13 |
| chr15_+_27739251 | 0.04 |
ENSRNOT00000011840
|
Parp2
|
poly (ADP-ribose) polymerase 2 |
| chr11_+_84396033 | 0.04 |
ENSRNOT00000002316
|
Abcc5
|
ATP binding cassette subfamily C member 5 |
| chr13_+_83721300 | 0.04 |
ENSRNOT00000082677
|
Adcy10
|
adenylate cyclase 10 (soluble) |
| chr2_-_123851854 | 0.04 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr11_-_31892531 | 0.04 |
ENSRNOT00000033745
|
Cryzl1
|
crystallin zeta like 1 |
| chr6_-_135049728 | 0.04 |
ENSRNOT00000009556
|
Hsp90aa1
|
heat shock protein 90 alpha family class A member 1 |
| chr8_-_109560747 | 0.04 |
ENSRNOT00000087334
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr6_-_36940868 | 0.04 |
ENSRNOT00000006470
|
Gen1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr4_-_133131953 | 0.04 |
ENSRNOT00000089177
|
Shq1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr14_-_17582823 | 0.04 |
ENSRNOT00000087686
|
LOC100911949
|
uncharacterized LOC100911949 |
| chr18_+_86299463 | 0.04 |
ENSRNOT00000058152
|
Cd226
|
CD226 molecule |
| chr9_+_9704346 | 0.04 |
ENSRNOT00000075403
|
Gpr108
|
G protein-coupled receptor 108 |
| chr17_-_78812111 | 0.04 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr1_+_80954858 | 0.04 |
ENSRNOT00000030440
|
Zfp112
|
zinc finger protein 112 |
| chr1_+_205706468 | 0.03 |
ENSRNOT00000089957
ENSRNOT00000023877 |
Edrf1
|
erythroid differentiation regulatory factor 1 |
| chr7_+_53878610 | 0.03 |
ENSRNOT00000091910
|
Osbpl8
|
oxysterol binding protein-like 8 |
| chr2_+_11658568 | 0.03 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr1_+_190462327 | 0.03 |
ENSRNOT00000030732
|
LOC691519
|
similar to ankyrin repeat domain 26 |
| chr13_-_47397890 | 0.03 |
ENSRNOT00000005505
|
C4bpb
|
complement component 4 binding protein, beta |
| chr3_+_130008885 | 0.03 |
ENSRNOT00000070972
|
Slx4ip
|
SLX4 interacting protein |
| chr2_-_54963448 | 0.03 |
ENSRNOT00000017886
|
Ptger4
|
prostaglandin E receptor 4 |
| chr12_+_24148567 | 0.03 |
ENSRNOT00000077378
ENSRNOT00000045157 |
Ccl24
|
C-C motif chemokine ligand 24 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat4_Stat3_Stat5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.2 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 0.0 | 0.3 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:2000814 | positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0042509 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.3 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.0 | GO:1903796 | negative regulation of inorganic anion transmembrane transport(GO:1903796) |
| 0.0 | 0.1 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0032197 | transposition, RNA-mediated(GO:0032197) |
| 0.0 | 0.1 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.0 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.1 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.0 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.0 | GO:0060545 | positive regulation of necroptotic process(GO:0060545) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.0 | GO:0046968 | cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 0.0 | 0.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.0 | GO:0002876 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.0 | 0.0 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.0 | 0.0 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.0 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.0 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.0 | GO:0090249 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 0.0 | 0.0 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.0 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.0 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.0 | GO:2000853 | negative regulation of corticosterone secretion(GO:2000853) |
| 0.0 | 0.0 | GO:1904976 | cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.0 | GO:0002424 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.0 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.0 | 0.1 | GO:1900115 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.0 | 0.0 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0098982 | glycine-gated chloride channel complex(GO:0016935) GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.0 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 0.3 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.4 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.0 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 0.0 | 0.0 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.0 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.0 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.0 | GO:0002134 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.0 | GO:2001070 | starch binding(GO:2001070) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |