Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
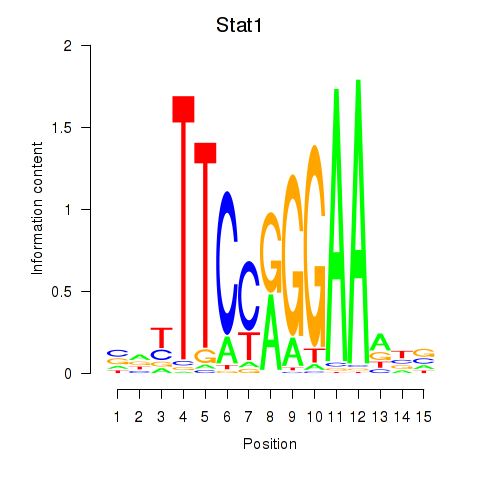
Results for Stat1
Z-value: 0.36
Transcription factors associated with Stat1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat1
|
ENSRNOG00000014079 | signal transducer and activator of transcription 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat1 | rn6_v1_chr9_-_54484533_54484533 | -0.73 | 1.6e-01 | Click! |
Activity profile of Stat1 motif
Sorted Z-values of Stat1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_37528011 | 0.27 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr20_-_3793985 | 0.20 |
ENSRNOT00000049540
ENSRNOT00000086293 |
RT1-CE16
|
RT1 class I, locus CE16 |
| chr7_-_3342491 | 0.17 |
ENSRNOT00000081756
|
Rdh5
|
retinol dehydrogenase 5 |
| chr2_+_145174876 | 0.15 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr9_+_15166118 | 0.10 |
ENSRNOT00000020432
|
Mdfi
|
MyoD family inhibitor |
| chr5_+_29538380 | 0.10 |
ENSRNOT00000010845
|
Calb1
|
calbindin 1 |
| chr10_+_84167331 | 0.09 |
ENSRNOT00000010965
|
Hoxb4
|
homeo box B4 |
| chr7_+_72924799 | 0.07 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr5_+_139597731 | 0.07 |
ENSRNOT00000072427
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr1_+_247519939 | 0.07 |
ENSRNOT00000034421
|
Cd274
|
CD274 molecule |
| chr2_+_114386019 | 0.06 |
ENSRNOT00000082148
|
AABR07009834.1
|
|
| chr1_-_81152384 | 0.06 |
ENSRNOT00000026263
ENSRNOT00000077305 |
Zfp94
|
zinc finger protein 94 |
| chr4_-_123494742 | 0.05 |
ENSRNOT00000073268
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr7_-_15163866 | 0.05 |
ENSRNOT00000042638
|
Zfp870
|
zinc finger protein 870 |
| chr6_+_108285822 | 0.05 |
ENSRNOT00000015889
|
Vsx2
|
visual system homeobox 2 |
| chr1_-_81412251 | 0.05 |
ENSRNOT00000026946
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr7_-_94563001 | 0.04 |
ENSRNOT00000051139
ENSRNOT00000005561 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr8_-_116349896 | 0.04 |
ENSRNOT00000087306
|
Lsmem2
|
leucine-rich single-pass membrane protein 2 |
| chr8_-_36764422 | 0.04 |
ENSRNOT00000017330
|
Hyls1
|
HYLS1, centriolar and ciliogenesis associated |
| chr1_-_98493978 | 0.04 |
ENSRNOT00000023942
|
Nkg7
|
natural killer cell granule protein 7 |
| chr3_+_95715193 | 0.04 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr1_+_87790104 | 0.04 |
ENSRNOT00000074580
|
LOC103690114
|
zinc finger protein 383-like |
| chr7_-_122644054 | 0.04 |
ENSRNOT00000025941
|
Dnajb7
|
DnaJ heat shock protein family (Hsp40) member B7 |
| chr11_+_80255790 | 0.04 |
ENSRNOT00000002522
|
Bcl6
|
B-cell CLL/lymphoma 6 |
| chr3_-_160139947 | 0.03 |
ENSRNOT00000014151
|
Ada
|
adenosine deaminase |
| chrX_+_13441558 | 0.03 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chr7_-_140090922 | 0.03 |
ENSRNOT00000014456
|
Lalba
|
lactalbumin, alpha |
| chr2_+_225827504 | 0.03 |
ENSRNOT00000018343
|
Gclm
|
glutamate cysteine ligase, modifier subunit |
| chr8_-_130429132 | 0.03 |
ENSRNOT00000026261
|
Hhatl
|
hedgehog acyltransferase-like |
| chr7_-_137856485 | 0.03 |
ENSRNOT00000007003
|
LOC688906
|
similar to splicing factor, arginine/serine-rich 2, interacting protein |
| chr13_-_45068077 | 0.03 |
ENSRNOT00000004969
|
Mcm6
|
minichromosome maintenance complex component 6 |
| chr2_-_165600748 | 0.03 |
ENSRNOT00000013216
|
Ift80
|
intraflagellar transport 80 |
| chr17_-_15467320 | 0.03 |
ENSRNOT00000072490
ENSRNOT00000093561 |
Ogn
|
osteoglycin |
| chr7_+_2718700 | 0.03 |
ENSRNOT00000050792
|
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr1_-_167093560 | 0.03 |
ENSRNOT00000027301
|
Il18bp
|
interleukin 18 binding protein |
| chr14_-_86387606 | 0.03 |
ENSRNOT00000089384
ENSRNOT00000006642 |
Ddx56
|
DEAD-box helicase 56 |
| chr3_-_112084144 | 0.03 |
ENSRNOT00000010959
|
Pla2g4f
|
phospholipase A2, group IVF |
| chr10_+_39109522 | 0.02 |
ENSRNOT00000010968
|
Irf1
|
interferon regulatory factor 1 |
| chr18_+_86299463 | 0.02 |
ENSRNOT00000058152
|
Cd226
|
CD226 molecule |
| chr17_+_18358712 | 0.02 |
ENSRNOT00000001979
|
Nup153
|
nucleoporin 153 |
| chr1_-_78180216 | 0.02 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr19_-_43215281 | 0.02 |
ENSRNOT00000025052
|
Aars
|
alanyl-tRNA synthetase |
| chr11_+_31806618 | 0.02 |
ENSRNOT00000043410
ENSRNOT00000002769 |
Son
|
Son DNA binding protein |
| chr19_-_43215077 | 0.02 |
ENSRNOT00000082151
|
Aars
|
alanyl-tRNA synthetase |
| chr1_-_189181901 | 0.02 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr2_+_30685840 | 0.02 |
ENSRNOT00000031385
|
Ccdc125
|
coiled-coil domain containing 125 |
| chr7_-_145174771 | 0.02 |
ENSRNOT00000055270
|
Glycam1
|
glycosylation dependent cell adhesion molecule 1 |
| chr4_+_109477920 | 0.02 |
ENSRNOT00000008468
|
Reg3a
|
regenerating islet-derived 3 alpha |
| chrX_+_156854594 | 0.02 |
ENSRNOT00000083442
|
Renbp
|
renin binding protein |
| chr15_+_33004133 | 0.02 |
ENSRNOT00000013246
|
Oxa1l
|
OXA1L, mitochondrial inner membrane protein |
| chr15_+_34282936 | 0.02 |
ENSRNOT00000026364
|
Irf9
|
interferon regulatory factor 9 |
| chr4_-_17594598 | 0.02 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr5_+_101526551 | 0.02 |
ENSRNOT00000015254
|
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr3_-_111873974 | 0.02 |
ENSRNOT00000077546
|
AABR07053509.2
|
|
| chr7_+_35125424 | 0.02 |
ENSRNOT00000085978
ENSRNOT00000010117 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chrX_-_123092217 | 0.02 |
ENSRNOT00000039710
|
Gm14569
|
predicted gene 14569 |
| chr10_+_17542374 | 0.02 |
ENSRNOT00000064079
|
Fbxw11
|
F-box and WD repeat domain containing 11 |
| chr18_+_36829062 | 0.02 |
ENSRNOT00000025467
ENSRNOT00000086979 |
Tcerg1
|
transcription elongation regulator 1 |
| chr4_+_118243132 | 0.02 |
ENSRNOT00000023654
|
RGD1306746
|
similar to Hypothetical protein MGC25529 |
| chr2_+_53109684 | 0.02 |
ENSRNOT00000086590
|
Selenop
|
selenoprotein P |
| chr17_+_47721977 | 0.02 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr4_+_9981958 | 0.02 |
ENSRNOT00000015322
|
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr15_+_34452116 | 0.02 |
ENSRNOT00000027647
|
Ltb4r
|
leukotriene B4 receptor |
| chr1_+_154131926 | 0.02 |
ENSRNOT00000035257
|
LOC102549852
|
ferritin light chain 1-like |
| chr1_+_214123044 | 0.02 |
ENSRNOT00000021925
|
Ptdss2
|
phosphatidylserine synthase 2 |
| chr6_+_109939345 | 0.02 |
ENSRNOT00000013560
|
Ift43
|
intraflagellar transport 43 |
| chr9_-_61134963 | 0.01 |
ENSRNOT00000017967
|
Pgap1
|
post-GPI attachment to proteins 1 |
| chr10_-_56506446 | 0.01 |
ENSRNOT00000021357
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr8_-_48850671 | 0.01 |
ENSRNOT00000016580
|
Cxcr5
|
C-X-C motif chemokine receptor 5 |
| chr15_-_34392066 | 0.01 |
ENSRNOT00000027315
|
Tgm1
|
transglutaminase 1 |
| chr10_-_59883839 | 0.01 |
ENSRNOT00000093579
|
Aspa
|
aspartoacylase |
| chr1_-_164441167 | 0.01 |
ENSRNOT00000023935
|
Rps3
|
ribosomal protein S3 |
| chrX_+_77065397 | 0.01 |
ENSRNOT00000090007
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr18_+_14471213 | 0.01 |
ENSRNOT00000045224
|
Dtna
|
dystrobrevin, alpha |
| chr5_-_56567320 | 0.01 |
ENSRNOT00000066081
|
Topors
|
TOP1 binding arginine/serine rich protein |
| chr19_+_37282018 | 0.01 |
ENSRNOT00000021723
|
Tmem208
|
transmembrane protein 208 |
| chr7_-_83348487 | 0.01 |
ENSRNOT00000006685
|
Nudcd1
|
NudC domain containing 1 |
| chr8_-_62332115 | 0.01 |
ENSRNOT00000025783
|
Mpi
|
mannose phosphate isomerase (mapped) |
| chrX_-_25590048 | 0.01 |
ENSRNOT00000004873
|
Mid1
|
midline 1 |
| chr8_+_21890319 | 0.01 |
ENSRNOT00000027949
|
LOC500956
|
hypothetical protein LOC500956 |
| chr10_+_14240219 | 0.01 |
ENSRNOT00000020233
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr13_+_90184569 | 0.01 |
ENSRNOT00000082393
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr5_-_169017295 | 0.01 |
ENSRNOT00000067481
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr5_+_164951789 | 0.01 |
ENSRNOT00000055658
ENSRNOT00000012193 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr19_+_15294248 | 0.01 |
ENSRNOT00000024622
|
Ces1f
|
carboxylesterase 1F |
| chr14_+_17195014 | 0.01 |
ENSRNOT00000031667
|
Cxcl11
|
C-X-C motif chemokine ligand 11 |
| chr14_-_23604834 | 0.01 |
ENSRNOT00000002760
|
Stap1
|
signal transducing adaptor family member 1 |
| chr1_+_226947105 | 0.01 |
ENSRNOT00000028373
|
Prpf19
|
pre-mRNA processing factor 19 |
| chr1_-_213921208 | 0.01 |
ENSRNOT00000044393
|
Ano9
|
anoctamin 9 |
| chr11_-_32088002 | 0.01 |
ENSRNOT00000002732
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr7_+_83348694 | 0.01 |
ENSRNOT00000006402
|
Eny2
|
ENY2, transcription and export complex 2 subunit |
| chr2_+_93518806 | 0.01 |
ENSRNOT00000050987
|
Snx16
|
sorting nexin 16 |
| chr11_+_68105369 | 0.01 |
ENSRNOT00000046888
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chrX_-_143453612 | 0.01 |
ENSRNOT00000051319
|
Atp11c
|
ATPase phospholipid transporting 11C |
| chr1_-_165606375 | 0.01 |
ENSRNOT00000024290
|
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr2_-_189880743 | 0.01 |
ENSRNOT00000018830
|
Snapin
|
SNAP-associated protein |
| chr19_-_37281933 | 0.01 |
ENSRNOT00000059663
|
Lrrc29
|
leucine rich repeat containing 29 |
| chr18_+_57654290 | 0.01 |
ENSRNOT00000025892
|
Htr4
|
5-hydroxytryptamine receptor 4 |
| chr3_+_176985900 | 0.01 |
ENSRNOT00000020210
|
Abhd16b
|
abhydrolase domain containing 16B |
| chr8_-_77992621 | 0.01 |
ENSRNOT00000085843
|
Myzap
|
myocardial zonula adherens protein |
| chr8_+_49621568 | 0.01 |
ENSRNOT00000021949
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr1_-_193700673 | 0.01 |
ENSRNOT00000071314
|
Paip2l1
|
polyadenylate-binding protein-interacting protein 2-like 1 |
| chr14_-_42560174 | 0.01 |
ENSRNOT00000003128
|
Tmem33
|
transmembrane protein 33 |
| chr13_-_47397890 | 0.01 |
ENSRNOT00000005505
|
C4bpb
|
complement component 4 binding protein, beta |
| chrX_-_38468360 | 0.01 |
ENSRNOT00000045285
|
Map7d2
|
MAP7 domain containing 2 |
| chr1_-_7480825 | 0.01 |
ENSRNOT00000048754
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr10_-_86645529 | 0.01 |
ENSRNOT00000011947
|
Med24
|
mediator complex subunit 24 |
| chr2_+_86996497 | 0.00 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr12_-_30106889 | 0.00 |
ENSRNOT00000065868
ENSRNOT00000001200 |
Tpst1
|
tyrosylprotein sulfotransferase 1 |
| chrX_+_9436707 | 0.00 |
ENSRNOT00000004187
|
Cask
|
calcium/calmodulin dependent serine protein kinase |
| chr3_+_161519743 | 0.00 |
ENSRNOT00000055148
|
Cd40
|
CD40 molecule |
| chrX_+_112270986 | 0.00 |
ENSRNOT00000091441
ENSRNOT00000082652 |
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr11_-_82938357 | 0.00 |
ENSRNOT00000035945
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr19_-_10884047 | 0.00 |
ENSRNOT00000088196
ENSRNOT00000063791 |
Cpne2
|
copine 2 |
| chr10_-_95262024 | 0.00 |
ENSRNOT00000066487
|
Bptf
|
bromodomain PHD finger transcription factor |
| chr7_-_2825498 | 0.00 |
ENSRNOT00000086656
ENSRNOT00000031362 |
Nabp2
|
nucleic acid binding protein 2 |
| chr10_+_69412017 | 0.00 |
ENSRNOT00000009448
|
Ccl2
|
C-C motif chemokine ligand 2 |
| chr7_+_1206648 | 0.00 |
ENSRNOT00000073689
|
Pros1
|
protein S (alpha) |
| chr15_-_103927592 | 0.00 |
ENSRNOT00000031036
ENSRNOT00000013366 |
Abcc4
|
ATP binding cassette subfamily C member 4 |
| chr20_+_45458558 | 0.00 |
ENSRNOT00000000713
|
Cdk19
|
cyclin-dependent kinase 19 |
| chr1_+_157403595 | 0.00 |
ENSRNOT00000012781
|
Ccdc90b
|
coiled-coil domain containing 90B |
| chr10_-_95431312 | 0.00 |
ENSRNOT00000085552
|
AABR07030603.2
|
|
| chr2_+_243820661 | 0.00 |
ENSRNOT00000090526
|
Eif4e
|
eukaryotic translation initiation factor 4E |
| chr7_-_102298522 | 0.00 |
ENSRNOT00000006273
|
A1bg
|
alpha-1-B glycoprotein |
| chr9_+_10952374 | 0.00 |
ENSRNOT00000074993
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chrX_+_159703578 | 0.00 |
ENSRNOT00000001162
|
Cd40lg
|
CD40 ligand |
| chr3_-_72171078 | 0.00 |
ENSRNOT00000009817
|
Serping1
|
serpin family G member 1 |
| chr2_+_199162745 | 0.00 |
ENSRNOT00000023488
|
Gja5
|
gap junction protein, alpha 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.0 | GO:1903165 | response to polycyclic arene(GO:1903165) |
| 0.0 | 0.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.0 | GO:0003322 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.0 | 0.0 | GO:0046103 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) inosine biosynthetic process(GO:0046103) negative regulation of mucus secretion(GO:0070256) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0099510 | calcium ion binding involved in regulation of cytosolic calcium ion concentration(GO:0099510) |
| 0.0 | 0.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |